Abstract
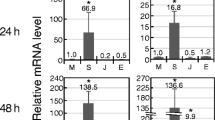
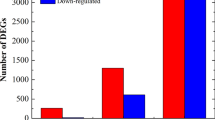
Ginger (Zingiber officinale Roscoe) cultivars are susceptible to soft rot disease caused by Pythium aphanidermatum. We analyzed changes in transcript levels of 41 genes in the highly susceptible ginger cultivar varada, a less susceptible wild accession (wild ginger), and a Pythium aphanidermatum-resistant relative, Z. zerumbet, following treatment with Pythium aphanidermatum or one of three signaling molecules: salicylic acid (SA), jasmonic acid (JA), or ethylene (ET). The 41 studied genes were chosen because they are known to be involved in the hypersensitive response (HR), cell signaling, or host defense. Expression of most genes peaked within 24 h of Pythium aphanidermatum infection. Interestingly, the level of induction was typically manyfold higher in Z. zerumbet than in wild ginger. However, several HR genes that were significantly induced in wild ginger were not induced in Z. zerumbet. Most of the genes, including those involved in signaling, did not respond to any of the three signaling molecules in Z. zerumbet while several genes responded to all the three signaling molecules in varada. In wild ginger, a large proportion of the genes responded to ET, but not to SA or JA. These results suggest that different mechanisms govern the three pathosystems. Resistance in Z. zerumbet seems to be independent of HR and the tested signaling molecules, whereas both mechanisms appear to be activated in the tolerance reaction of wild ginger. This work revealed potential defense components of this understudied tropical taxa, and will contribute to the design of strategies for transgenic improvement of ginger.


Similar content being viewed by others
References
Agrios GN (2004) Plant pathology, 5th edn. Academic, San Diego
Ameline-Torregrosa C, Dumas B, Krajinski F, Esquerre-Tugaye MT, Jacquet C (2006) Transcriptomic approaches to unravel plant-pathogen interactions in legumes. Euphytica 147:25–36
Aswati Nair R, Thomas G (2007) Isolation, characterization and expression studies of resistance gene candidates (RGCs) from Zingiber spp. Theor Appl Genet 116:123–134
Basse CW (2005) Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol 138:1774–1784
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Broekaert WF, Delaure SL, De Bolle MFC, Cammue BPA (2006) The role of ethylene in host–pathogen interactions. Annu Rev Phytopathol 44:393–416
Chen LR, Chen YJ, Lee CY, Lin TY (2007) MeJA-induced transcriptional changes in adventitious roots of Bupleurum kaoi. Plant Sci 173:12–24
Coram TE, Pang ECK (2006) Expression profiling of chickpea genes differentially regulated during a resistance response to Ascochyta rabiei. Plant Biotechnol J 4:647–666
Dake GN (1995) Diseases of ginger (Zingiber officinale Rosc.) and their management. J Spices Aromat Crops 4:40–48
Eichmann R, Biemelt S, Schafer P, Scholz U, Jansen C, Felk A, Schafer W, Langen G, Sonnewald U, Kogel KH, Huckelhoven R (2006) Macroarray expression analysis of barley susceptible and nonhost resistance to Blumeria graminis. J Plant Physiol 163:657–670
Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61:243–282
Folman LB, Postma J, van Veen JA (2003) Inability to find consistent bacterial biocontrol agents of Pythium aphanidermatum in cucumber using screens based on ecophysiological traits. Microbiol Ecol 45:72–87
Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98:373–378
Gong H, Hu WW, Pua EC (2005) The role of a Raf-related kinase gene from mustard (Brassica juncea) in glutathione-related signaling pathway. Plant Sci 169:255–265
Grover A, Gowthaman R (2003) Strategies for development of fungus-resistant transgenic plants. Curr Sci 84:330–340
Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7:40–49
Hajjar R, Hodgkin T (2007) The use of wild relatives in crop improvement: a survey of development over the last 25 years. Euphytica 156:1–13
Hammond-Kosack KE, Parker JE (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14:177–193
Jones DA, Takemoto D (2004) Plant innate immunity-direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immunol 16:48–62
Kamoun S, Huitema E, Vleeshouwers VGAA (1999) Resistance to oomycetes: a general role for the hypersensitive response? Trends Plant Sci 4:196–200
Kavitha PG (2006) Genetic variation in wild gingers (Zingiberaceae) for resistance to Pythium infection and analysis of differentially expressed transcripts in hosts with different levels of pathogen response. PhD thesis, University of Kerala
Kavitha PG, Thomas G (2007) Evaluation of Zingiberaceae for resistance to ginger soft rot caused by Pythium aphanidermatum (Edson) Fitzp. Plant Genet Resour Newsl 152:1–4
Kavitha PG, Thomas G (2008a) Population genetic structure of the clonal plant Zingiber zerumbet (L.) Smith (Zingiberaceae), a wild relative of cultivated ginger, and its response to Pythium aphanidermatum. Euphytica 160:89–100
Kavitha PG, Thomas G (2008b) Defence transcriptome profiling of Zingiber zerumbet (L.) Smith by mRNA differential display. J Biosci. doi:10.1007/s12038-008-0002-2
Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Mattews BF (2007) A time-course comparative microarray analysis of an imcompatible and compatible response by Glycine max (soybean) to Heterodera glycines (soybean cyst nematode) infection. Planta 226:1423–1447
Lawrence BM (1984) Major tropical spices-Ginger (Zingiber officinale Rosc.). Perfum Flavor 9:1–40
Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967–971
Ma QH, Wang XM (2003) Characterization of an ethylene receptor homologue from wheat and its expression during leaf senescence. J Exp Bot 54:1489–1490
Moy P, Qutob D, Chapman BP, Atkinson I, Gijzen M (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant Microbe Interact 17:1051–1062
Okubara PA, Paulitz TC (2005) Root defense responses to fungal pathogens: a molecular perspective. Plant Soil 274:215–226
Parlevliet JE (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124:147–156
Ravindran PN, Sasikumar B, George JK, Ratnambal MJ, Nirmal Babu K, Zachariah JT, Nair RR (1994) Genetic resources of ginger (Zingiber officinale Rosc.) and its conservation in India. Plant Genet Resour Newsl 98:1–4
Robb J, Lee B, Nazar RN (2007) Gene expression in a tolerant tomato–vascular pathogen interaction. Planta 226:299–309
Roetschi A, Si-Ammour A, Belbahri L, Mauch F, Mauch-Mani B (2001) Characterization of an Arabidopsis–Phytophthroa pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signaling. Plant J 28:293–305
Roy BA, Kirchner JW (2000) Evolutionary dynamics of pathogen resistance and tolerance. Evolution 54:51–63
Salzman RA, Fujita T, Zhu-Salzman K, Hasegawa PM, Bressan RA (1999) An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Mol Biol Rep 17:11–17
Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277:10555–10561
Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97:11655–11660
Selvan MT, Thomas KG, Manojkumar K (2002) Ginger (Zingiber officinale Rosc.). In: Singh HP, Sivaraman K, Selvan MT (eds) Indian spices—production and utilization. Coconut Development Board, Kochi, pp 110–131
Smart CD, Myers KL, Restrepo S, Martin GB, Fry WE (2003) Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid or salicylic acid signaling pathways. Mol Plant Microbe Interact 16:141–148
Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15:317–330
Tian ZD, Liu J, Wang BL, Xie CH (2006) Screening and expression analysis of Phytophthora infestans induced genes in potato leaves with horizontal resistance. Plant Cell Rep 25:1094–1103
Torregrosa C, Cluzet S, Fournier J, Huguet T, Gamas P, Prosperi JM, Esquerre-Tugaye MT, Dumas B, Jacquet C (2004) Cytological, genetic and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol Plant Microbe Interact 17:909–920
Torres GAM, Pflieger S, Corre-Menguy F, Mazubert C, Hartmann C, Lelandais-Briere C (2006) Identification of novel drought-related mRNAs in common bean roots by differential display RT-PCR. Plant Sci 171:300–307
Vaghchhipawala ZE, Schlueter JA, Shoemaker RC, Mackenzie SA (2004) Soybean FGAM synthase promoters direct ectopic nematode feeding site activity. Genome 47:404–413
van Kan JAL (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci 11:247–253
Vega-Sánchez ME, Redinbaugh MG, Costanzo S, Dorrance AE (2005) Spatial and temporal expression analysis of defense-related genes in soybean cultivars with different levels of partial resistance to Phytophthora sojae. Physiol Mol Plant Pathol 66:175–182
Wan J, Dunning FM, Bent AF (2002) Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays. Funct Integr Genomics 2:259–273
Wang B, Liu J, Tian Z, Song B, Xie C (2005) Monitoring the expression patterns of potato genes associated with quantitative resistance to late blight during Phytophthora infestans infection using cDNA microarrays. Plant Sci 169:1155–1167
Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PRJ (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450:115–119
Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA (2007) Transcript profiling in host–pathogen interactions. Annu Rev Phytopathol 45:329–369
Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, Zhao YX, Lu Y, Ni ZY, Liu L, Qiu ZG, Ma YZ (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress response. Plant Mol Biol 65:719–732
Zhao J, Wang J, An L, Doerge RW, Chen ZJ, Grau CR, Meng J, Osborn TC (2007) Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227:13–24
Zhou HL, Cao WH, Cao YR, Liu J, Hao YJ, Zhang JS, Chen SY (2006) Roles of ethylene receptor NTHK1 domains in plant growth, stress response and protein phosphorylation. FEBS Lett 580:1239–1250
Acknowledgments
P. G. Kavitha gratefully acknowledges the Council for Scientific and Industrial Research (CSIR), Government of India for the research fellowship (File No. 9/716(024)/2KI/EMR-I) and G. Thomas acknowledges the Department of Biotechnology (DBT), Government of India for the research grant (Grant No. BT/PR2211/Agr/08/162/2001). The authors thank the Indian Institute of Spices Research, Calicut, India for the fungal strain and Dr Sabu, University of Calicut, Kerala, India for the wild accessions of ginger.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Lakshmanan.
Rights and permissions
About this article
Cite this article
Kavitha, P.G., Thomas, G. Expression analysis of defense-related genes in Zingiber (Zingiberaceae) species with different levels of compatibility to the soft rot pathogen Pythium aphanidermatum . Plant Cell Rep 27, 1767–1776 (2008). https://doi.org/10.1007/s00299-008-0594-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0594-x