Abstract
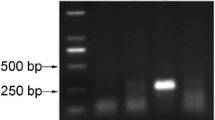
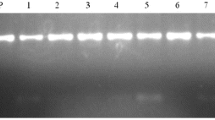
Citrus is an economically important fruit crop, belongs to family Rutaceae, cultivated commercially in over 130 countries, which holds a leading profitable position in the international market. The most important citrus varieties are mandarins, oranges, lemons, sweet limes, grapefruits and pomelos. Citrus yellow vein clearing virus (CYVCV) is an important graft transmissible plant pathogen known to reduce productivity of citrus fruits due to its predominant association and widespread occurrence. Requirement of fast, reliable, efficient & economical CYVCV indexing assay is a prerequisite for production of healthy planting material. Currently, nucleic acid isolation and thermal cycler-based assay available for CYVCV indexing is a cumbersome lab intensive method. The present study was undertaken to develop and validate reverse transcription-recombinase polymerase amplification (RT-RPA) assay requiring no tedious RNA isolation, separate cDNA synthesis and costlier instrument like thermo-cycler. Optimized RT-RPA assay was able to amplify CYVCV up to 10–7 dilution (equivalent to 0.1 pg/μl) with the prepared templates of both RNA and crude saps and showed higher sensitivity in detection of CYVCV infection in field samples as compared to the conventional RT-PCR. Developed RT-RPA assay showed high specificity without any cross-reaction with other citrus pathogens (Indian citrus ringspot virus, citrus yellow mosaic virus, citrus tristeza virus, citrus exocortis viroid and huanglongbing). RT-RPA using crude leaf sap as template is quite simple, robust, highly sensitive, time and cost effective; therefore, it can be used in resource constrained laboratories as screening tool, for field surveys and on-site testing programs in farms, nurseries and biosecurity. Present study, first time reports the development, optimization and validation of crude sap-based RT-RPA assay for the detection of CYVCV infection in citrus plants namely; Kinnow mandarin, Mosambi and Grape fruit.



Similar content being viewed by others
Data Availability
Sequence generated in this study was submitted in NCBI GenBank with accession number OQ427642.
References
Alshami AAA, Ahlawat YS, Pant RP (2003) A hitherto unreported yellow vein clearing disease of citrus in India and its viral etiology. Indian Phytopathol 56(4):422–427
Catara A, Azzaro A, Davino M, Polizzi G (1993) Yellow vein clearing of lemon in Pakistan. In: Proceedings of 12th conference IOCV, Riverside, CA, pp 364–367
Ahlawat YS, Pant RP (2003) Major viruses and virus-like diseases India, their diagnosis and management. Annu Rev Plant Pathol 2:447–474
Chen H, Li Z, Wang X, Zhou Y, Tang K, Zhou C, Zhao X, Yue J (2014) First report of citrus yellow vein clearing virus on lemon in Yunnan China. China Plant Dis 98:1747. https://doi.org/10.1094/PDIS-04-14-0343-PDN
Bani SMH, Aghajanzadeh S (2017) Occurrence of citrus yellow vein clearing virus in citrus species in Iran. J Plant Pathol 99(1):290
Zhou Y, Chen HM, Cao MJ, Wang XF, Jin X, Liu KH, Zhou CY (2017) Occurrence, distribution and molecular characterization of citrus yellow vein clearing virus in China. Plant Dis 101:137–143. https://doi.org/10.1094/PDIS-05-16-0679-RE
Onelge N, Satar S, Elibuyuk O, Bozan O, Kamberoolu M (2011) Transmission studies on citrus yellow vein clearing virus. Int Org Citrus Virol Conf Proceed 18:18
Song Z, Kurth EG, Peremyslov VV, Zhou CY (2015) Molecular characterization of a citrus yellow vein clearing virus strain from China. Arch Virol 160:1811–1813
Loconsole G, Önelge N, Potere O, Giampetruzzi A, Bozan O, Satar S, De Stradis A, Savino V, Yokomi RK, Saponari M (2012) Identification and characterization of citrus yellow vein clearing virus, a putative new member of the genus Mandarivirus. Phytopathology 102:1168–1175. https://doi.org/10.1094/PHYTO-06-12-0140-R
Rustici G, Milne RG, Accotto GP (2002) Nucleotide sequences, genome organization and phylogenetic analysis of Indian citrus ringspot virus. Arch Virol 147:2215–2224. https://doi.org/10.1007/s0070
Adams MJ, Antoniw JF, Bar-Joseph M, Brunt AA, Candresse T, Foster GD, Martelli GP, Milne RG, Zavriev SK, Fauquet CM (2004) The new plant virus family Flexi viridae and assessment of molecular criteria for species demarcation. Arch Virol 149:1045–1060
Adams MJ, Lefkowitz EJ, King AMQ, Carstens EB (2014) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014). Arch Virol 159:2831–2841. https://doi.org/10.1007/s0070
Zhen S, Kurth EG, Peremysolv VV, Changyong Z, Dolja VV (2015) Molecular characterization of a citrus yellow vein clearing virus strain from China. Arch Virol 160(7):1811–1813. https://doi.org/10.1007/s00705-015-2423-1
Meena RP, Prabha K, Baranwal VK (2019) Genome characterization of citrus yellow vein-clearing virus: limited heterogeneity of viral genomes in Mandarivirus-infecting different citrus species. 3 Biotech 9:1–1
Liu Z, Sun YJ, Zhou XP, Hong J, Wu JX (2017) Monoclonal antibody-based serological detection of citrus yellow vein clearing virus in citrus groves. J Integr Agr 16:884–891
Kokane AD, Lawrence K, Kokane SB, Gubyad MG, Misra P, Reddy MK, Ghosh DK (2021) Development of a SYBR Green-based RT-qPCR assay for the detection of Indian citrus ringspot virus. 3 Biotech 11:1–2
Sharma S (2022) Molecular indexing against Mandariviruses and Citrus greening bacterium in Kinnow mandarin nurseries in Punjab. Ind Phytopathol 75(3):895–899
Penchalaiah Y, Gopi V, Gopal K, Baranwal VK (2008) Detection of citrus ring spot virus and citrus yellow vein clearing virus in sweet orange by DAC-ELISA. Ann Plant Protecti Sci 16:153–155
Bin Y, Song Z, Li ZA, Zhou CY (2015) Direct tissue blot immunoassay for detection of citrus yellow vein clearing virus. Acta Hortic Sin 42:1843–1850
Bin Y, Li ZA, Wu JX, Wang XF, Zhou Y, Li TS, Yang FY, Zhou CY, Song Z (2018) Development of an immune-chromatographic strip test for rapid detection of citrus yellow vein clearing virus. Arch Virol 163:349–357
Meena RP, Baranwal VK (2016) Development of multiplex polymerase chain reaction assay for simultaneous detection of clostero-, badna- and mandari-viruses along with huanglongbing bacterium in citrus trees. J Virol Methods 235:58–64. https://doi.org/10.1016/j.jviromet.2016.05.012
Chen HM, Zhou Y, Wang XF, Zhou CY, Yang XY, Li ZA (2016) Detection of citrus yellow vein clearing virus based on a real time RT-PCR approach. Acta Hortic Sin 43:168–174
Liu KL, Chen HM, Zhou Y, Li ZA (2015) Establishment of RT-LAMP assay for detection of citrus yellow vein clearing virus. Acta Horti Sin 42:997–1002
Zhou Y, Chen HM, Wang XF, Li ZA, Wang L, Zhou CY (2016) Development and application of nested RT-PCR assay for detection of citrus yellow vein clearing virus. J Plant Protect 43:255–259
Piepenburg O, Williams CH, Stemple DL, Armes NA (2006) DNA detection using recombination proteins. PLoS Biol 4(7):1115–1121
Daher RK, Stewart G, Boissinot M, Bergeron MG (2016) Recombinase polymerase amplification for diagnostic applications. Clin Chem 62(7):947–958
Kapoor R, Srivastava N, Kumar S, Saritha RK, Sharma SK, Jain RK, Baranwal VK (2017) Development of a recombinase polymerase amplification assay for the diagnosis of Banana bunchy top virus in different banana cultivars. Arch Virol 162:2791–2796
Kumar PV, Sharma SK, Rishi N, Ghosh DK, Baranwal VK (2018) An isothermal based recombinase polymerase amplification assay for rapid, sensitive, and robust indexing of citrus yellow mosaic virus. Acta Virol 62:104–108
Srivastava N, Kapoor R, Kumar R, Kumar S, Saritha RK, Kumar S, Baranwal VK (2019) Rapid diagnosis of Cucumber mosaic virus in banana plants using a fluorescence-based real-time isothermal reverse transcription-recombinase polymerase amplification assay. J Virol Methods 270:52–58
Kumar R, Pant RP, Kapoor S, Khar A, Baranwal VK (2021) Development of a reverse transcription-recombinase polymerase amplification (RT-RPA) assay for the detection of onion yellow dwarf virus (OYDV) in onion cultivars. Indian Phytopathol 74:201–207
Sharma SK, Pathaw N, Wangkhem B, Jackson KS, Devi KS, Roy SS, Singh AR, Singh R, Banerjee A, Kumar S, Ningombam A (2022) Simple template-based reverse transcription-recombinase polymerase amplification assay for routine diagnosis of citrus tristeza virus. Lett Appl Microbiol 76:1–9. https://doi.org/10.1093/lambio/ovac060
Kishan G, Kumar R, Sharma SK, Srivastava N, Gupta N, Kumar A, Baranwal VK (2023) Development and application of crude sap-based recombinase polymerase amplification assay for the detection and occurrence of grapevine geminivirus a in Indian Grapevine cultivars. Front Plant Sci. https://doi.org/10.3389/fpls.2023.1151471
Chen J (2010) Molecular detection of Cucumber Mosaic Virus and other RNA viruses based on new techniques. Experimental plant virology. Advanced topics in science and technology in China. Springer, Berlin
Mekuria TA, Zhang S, Eastwel KC (2014) Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification. J Virol Methods 205:24–30
Zhang S, Ravelonandro M, Russell P, Mc Owen N, Briard P, Bohannon S, Vrient A (2014) Rapid diagnostic detection of plum poxvirus in Prunus plants by isothermal AmplifyRP® using reverse transcription-recombinase polymerase amplification. J Virol Methods 207:114–120
Silva G, Bömer M, Nkere C, Kumar PL, Seal SE (2015) Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J Virol Methods 222:138–144
Babu B, Washburn BK, Miller SH, Poduch K, Sarigul T, Knox GW, Ochoa-Corona FM, Paret ML (2017) A rapid assay for detection of rose rosette virus using reverse transcription-recombinase polymerase amplification using multiple gene targets. J Virol Methods 240:78–84
Jiao Y, Xu C, Li J, Gu Y, Xia C, Xie Q, Xie Y, An M, Xia Z, Wu Y (2020) Characterization and a RT-RPA assay for rapid detection of chilli veinal mottle virus (ChiVMV) in tobacco. Virol J 17:33
Sambrook J, Russell DW (2006) The condensed protocols from molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbour Laboratory Cold Spring Harbour Press, New York
Liu Y, Wang Y, Wang Q, Zhang Y, Shen W, Li R, Cao M, Chen L, Li X, Zhou C, Zhou Y (2019) Development of a sensitive and reliable reverse transcription droplet digital PCR assay for the detection of citrus yellow vein clearing virus. Arch Virol 164:691–697
Bakheit MA, Torra D, Palomino LA, Thekisoe OM, Mbati PA, Ongerth J, Karanis P (2008) Sensitive and specific detection of Cryptosporidium species in PCR-negative samples by loop-mediated isothermal DNA amplification and confirmation of generated LAMP products by sequencing. Vet Parasitol 158(1–2):11–22
Londono MA, Harmon CL, Polston JE (2016) Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol J 13:48
Tomar S, Lavickova B, Guiducci C (2022) Recombinase polymerase amplification in minimally buffered conditions. Biosens Bioelectron 15(198):113802
Devi HC, Devi KS, Sanabam R, Pathaw N, Devi OP, Chanu NT, Maibam A, Chanu WT, Sanasam J, Roy SS, Devi CP, Singh AR, Devi PS, Gupta N, Sharma SK (2023) Simplified extraction protocol for plant tissues and reverse transcription RPA assay for quick and reliable diagnosis and its application in resistance screening of chilli veinal mottle virus. Crop Protect 1(170):106280
Acknowledgements
The authors wish to thank the Head of the Division of Plant Pathology and Director, ICAR-Indian Agricultural Research Institute, New Delhi, India for laboratory facilities and encouragement.
Funding
This study was funded by ICAR-National Professor Project entitled “Virome analysis of vegetatively propagated fruit crops for enhancing the productivity through virus prevention program” (Ag.Edn.F.No./27/01/NP/2022-HRD) and Department of Biotechnology, Government of India project entitled “National Certification System for Tissue Culture Raised Plants (NCS-TCP)’ (NO. BT/AB/03/02/2021).
Author information
Authors and Affiliations
Contributions
Virendra Kumar Baranwal and Nitika Gupta supervised and conceptualized the whole research project. Nitika Gupta and Susheel Kumar Sharma edited the manuscript. Rakesh Kumar conducted the experiments, Nitika Gupta and Rakesh Kumar have written the manuscript. Gopi Kishan has supported in conduction of lab experiments and manuscript writing. Ashwini Kumar and Nishant Srivastva contributed in survey and sample collection.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gupta, N., Kumar, R., Kishan, G. et al. Development of Simplified Recombinase Polymerase Amplification Assay for Rapid and Robust Detection of Citrus Yellow Vein Clearing Virus. Curr Microbiol 81, 103 (2024). https://doi.org/10.1007/s00284-024-03614-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-024-03614-y