Abstract
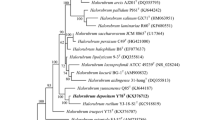
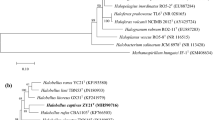
An extremely halophilic archaeon strain named FL173T was isolated from a salt mine (Anhui Province, China). Colonies on agar plate are orange-red, moist, and opaque. Cells are motile, Gram-stain-negative, polymorphic, and lyse in distilled water. Cells are able to grow at temperatures, NaCl concentrations, and pH ranging from 20 to 50 °C (optimum 42 °C), 2.6 to 5.1 M NaCl concentration (optimum 3.4 M), and 5.5 to 9.5 pH (optimum 7.0), respectively. Mg2+ is not necessary for growth. The major polar lipids of strain FL173T were phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfonate (PGS), sulfonated mannosyl glycolipid (S-DGD-1). It has two copies of the 16S rRNA gene, which share the highest sequence similarity (93.04–99.02% sequence similarity) to the 16S rRNA genes of Halomicroarcula salinisoli F24AT, respectively. The rpoB' gene of strain FL173T showed the highest sequence similarity (93.76%) to that of H. salinisoli F24AT. The genome-based analysis showed that the average amino-acid identity (AAI), orthologous average nucleotide identity (ANI) and in silico DNA–DNA hybridization values between strains FL173T and H. salinisoli F24AT were 84.80%, 85.29%, and 29.70%, respectively, which are far below the threshold for the delineation of a prokaryotic new species. The DNA G+C content of strain FL173T is 64.9%. Genomic, physiological, biochemical, and phenotypic evidences showed that strain FL173T (CGMCC 1.18851=NBRC 114260) represents a new species of the genus Halomicroarcula, for which the name Halomicroarcula salaria sp. nov. is proposed.



Similar content being viewed by others
Abbreviations
- AAI:
-
Average amino-acid identity
- ANI:
-
Average nucleotide identity
- DDH:
-
DNA–DNA hybridization
- BLAST:
-
Basic Local Alignment Search Tool
- ML:
-
Maximum-Likelihood
- MP:
-
Maximum-Parsimony
- NJ:
-
Neighbor-Joining
- DMSO:
-
Dimethylsulfoxide
- PG:
-
Phosphatidylglycerol\
- PGP-Me:
-
Phosphatidylglycerol phosphate methyl ester
- PGS:
-
Phosphatidylglycerol sulfate
- S-DGD-1:
-
Sulfated mannosyl glucosyl diether
- GL:
-
Glycolipid
- TLC:
-
Thin-layer chromatography
- CGMCC:
-
China General Microbiological Culture Collection Center
- MES:
-
2-Morpholinoethanesulfonic acid
- PIPES:
-
1, 4-Piperazine bis-ethanesulfonic acid
- CHES:
-
2-Cyclohexylamino ethanesulfonic acid
- CAPS:
-
N-Cyclohexyl-3-aminopropanesulfonic acid
References
Ventosa A (N.D.) Unusual microorganisms from unusual habitats: hypersaline environments. In: Prokaryotic diversity—mechanisms and significance. Society for General Microbiology symposium no. 66. https://doi.org/10.1017/cbo9780511754913.015
Ventosa A, de la Haba RR, Sánchez-Porro C, Papke RT (2015) Microbial diversity of hypersaline environments: a metagenomic approach. Curr Opin Microbiol 25:80–87. https://doi.org/10.1016/j.mib.2015.05.002
Loukas A, Kappas I, Abatzopoulos TJ (2018) HaloDom: a new database of halophiles across all life domains. J Biol Res (Thessalon) 25:2. https://doi.org/10.1186/s40709-017-0072-0
Echigo A, Minegishi H, Shimane Y, Kamekura M, Itoh T, Usami R (2013) Halomicroarcula pellucida gen. nov., sp. nov., a non-pigmented, transparent-colony-forming, halophilic archaeon isolated from solar salt. Int J Syst Evol Microbiol 63:3556–3562. https://doi.org/10.1099/ijs.0.049965-0
Zhang WJ, Cui HL (2014) Halomicroarcula limicola sp. nov., isolated from a marine solar saltern, and emended description of the genus Halomicroarcula. Int J Syst Evol Microbiol 64:1747–1751. https://doi.org/10.1099/ijs.0.062455-0
Chen F, Xu Y, Sun S, Shi X, Liu A, Chen S (2020) Halomicroarcula amylolytica sp. nov., a novel halophilic archaeon isolated from a salt mine. Int J Syst Evol Microbiol 70:4978–4985. https://doi.org/10.1099/ijsem.0.004368
Zhang WJ, Cui HL (2015) Halomicroarcula salina sp. nov., isolated from a marine solar saltern. Int J Syst Evol Microbiol 65:1628–1633. https://doi.org/10.1099/ijs.0.000150
Durán-Viseras A, Sánchez-Porro C, Ventosa A (2021) Genomic insights into new species of the genus Halomicroarcula reveals potential for new osmoadaptative strategies in halophilic archaea. Front Microbiol 12:751746. https://doi.org/10.3389/fmicb.2021.751746
Tu D, Ke J, Luo Y, Hong T, Sun S, Han J, Chen S (2022) Microbial community structure and shift pattern of industry brine after a long-term static storage in closed tank. Front Microbiol 13:975271. https://doi.org/10.3389/fmicb.2022.975271
Kharroub K, Quesada T, Ferrer R, Fuentes S, Aguilera M, Boulahrouf A, Ramos-Cormenzana A, Monteoliva-Sánchez M (2006) Halorubrum ezzemoulense sp. nov., a halophilic archaeon isolated from Ezzemoul Sabkha Algeria. Int J Syst Evol Microbiol 56:1583–1588. https://doi.org/10.1099/ijs.0.64272-0
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B’ (rpoB’) gene. Int J Syst Evol Microbiol 60:2398–2408. https://doi.org/10.1099/ijs.0.017160-0
Jin JQ, Sun YB (2018) AutoSeqMan: batch assembly of contigs for Sanger sequences. Zool Res 39:123–126. https://doi.org/10.24272/j.issn.2095-8137.2018.027
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser 41:95–98. https://doi.org/10.1021/bk-1999-0734.ch008
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Oren A, Ventosa A, Grent WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol. https://doi.org/10.1099/00207713-47-1-233
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485. https://doi.org/10.1128/jb.70.4.484-485.1955
Hartmann R, Sickinger HD, Oesterhelt D (1980) Anaerobic growth of halobacteria. Proc Natl Acad Sci USA 77:3821–3825. https://doi.org/10.1073/pnas.77.7.3821
Pai S-C, Yang C-C, Riley PJ (1990) Formation kinetics of the pink azo dye in the determination of nitrite in natural waters. Anal Chim Acta 232:345–349. https://doi.org/10.1016/S0003-2670(00)81252-0
Cui HL, Lin ZY, Dong Y, Zhou PJ, Liu SJ (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206. https://doi.org/10.1099/ijs.0.65268-0
Benson HJ (2002) Microbiological applications: laboratory manual in general microbiology. McGraw-Hill, New York
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715. https://doi.org/10.1139/m78-119
Holding AJ, Collee JG (1971) Chapter I routine biochemical tests. In: Norris JR, Ribbons DW (eds) Methods in microbiology. Academic, New York, pp 1–32. https://doi.org/10.1016/s0580-9517(08)70573-7
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. https://doi.org/10.1101/gr.186072.114
Kumar S, Bansal K, Sethi SK (2022) Reclassification of Streptococcus ilei as a later heterotypic synonym of Streptococcus koreensis based on whole-genome sequence analysis. Arch Microbiol 204:408. https://doi.org/10.1007/s00203-022-02979-7
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Medlar AJ, Törönen P, Holm L (2018) AAI-profiler: fast proteome-wide exploratory analysis reveals taxonomic identity, misclassification and contamination. Nucleic Acids Res 46:W479–W485. https://doi.org/10.1093/nar/gky359
Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP (2013) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418. https://doi.org/10.1007/s00203-013-0888-4
Xin YJ, Bao CX, Tan S, Hou J, Cui HL (2022) Haladaptatus halobius sp. nov. and Haladaptatus salinisoli sp. Nov., two extremely halophilic archaea isolated from Gobi saline soil. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.005543
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354-357. https://doi.org/10.1093/nar/gkj102
Xu L, Dong Z, Fang L et al (2019) OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 47(W1):W52–W58. https://doi.org/10.1093/nar/gkz333
Ali A, Tariq H, Abbas S, Arshad M, Li S, Dong L, Li L, Li WJ, Ahmed I (2021) Draft genome sequence of a multidrug-resistant novel candidate Pseudomonas sp. NCCP-436(T) isolated from faeces of a bovine host in Pakistan. J Glob Antimicrob Resist 27:91–94. https://doi.org/10.1016/j.jgar.2021.08.011
Corcelli A, Lobasso S (2006) 25 characterization of lipids of halophilic archaea. Methods Microbiol 35:585–613. https://doi.org/10.1016/s0580-9517(08)70028-x
Minnikin DE, Apos Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–242. https://doi.org/10.1016/0167-7012(84)90018-6
Stropko SJ, Pipes SE, Newman JD (2014) Genome-based reclassification of Bacillus cibi as a later heterotypic synonym of Bacillus indicus and emended description of Bacillus indicus. Int J Syst Evol Microbiol 64(Pt 11):3804–3809. https://doi.org/10.1099/ijs.0.068205-0
Acknowledgements
The authors gratefully acknowledge Mr. Feng Li from Dingyuan Salt Mine for assisting in sample collection, Mr. Hao Wu and Tingting Chen from Anhui Normal University for collation of the data.
Funding
This work was supported by Grants from the Anhui Provincial Key Laboratory of Conservation and Exploitation of Biological Resources (swzy202011), the Natural Science Foundation of Anhui Province (2208085MC39), the Opening Project of the State Key Laboratory of Microbial Resources (SKLMR-20220702), the Outstanding Innovative Research Team for Molecular Enzymology and Detection in Anhui Provincial Universities (2022AH010012), the Innovation and Entrepreneurship Training Program of Anhui Normal University for Undergraduates (Nos. S202210370295 and 202210370139).
Author information
Authors and Affiliations
Contributions
SC and PL: conceptualization and funding acquisition. TH, JK, LC and YH: data curation. TH and LC: investigation. TH and LC: methodology. TH and SC: writing-original draft. TH, JK and SC: writing-review and editing. All authors have read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hong, T., Ke, J., Chen, L. et al. Genomic, Physiological, Biochemical, and Phenotypic Evidences Reveal a New Species, Halomicroarcula salaria sp. nov. Curr Microbiol 81, 71 (2024). https://doi.org/10.1007/s00284-023-03574-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03574-9