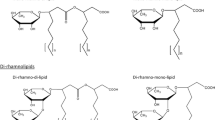
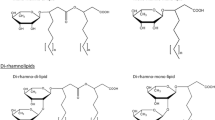
Abstract
Rhamnolipid congeners have been widely used in agriculture and biomedicine as potent surfactants. They have recently attracted attention due to their diverse and versatile biological functions, which include an important bacterial virulence factor that makes them attractive targets for research into biosynthetic pathways and gene regulation. The intricate gene expression and regulation network controlling their biosynthesis remain to be completely understood. This article summarizes current knowledge about the biosynthesis pathways and regulatory mechanisms of rhamnolipid congeners, that meet the pharmacological needs of human health and agriculture.



Similar content being viewed by others
References
Raza ZA, Khalid ZM, Banat IM (2009) Characterization of rhamnolipids produced by a Pseudomonas aeruginosa mutant strain grown on waste oils. J Environ Sci Heal 44:1367–1373. https://doi.org/10.1080/10934520903217138
Nakata K, Ishigami Y (1999) A facile procedure of remediation for oily waste with rhamnolipid biosurfactant. J Environ Sci Heal 34:1129–1142. https://doi.org/10.1080/10934529909376886
Chen J, Wu Q, Hua Y, Chen J, Zhang H, Wang H (2017) Potential applications of biosurfactant rhamnolipids in agriculture and biomedicine. Appl Microbiol Biotechnol 101:1–11. https://doi.org/10.1016/j.actamat.2017.07.029
Brenner DJ, McWhorter AC, Kai A, Steigerwalt AG, Farmer JJ (1986) Enterobacter asburiae sp. nov., a new species found in clinical specimens, and reassignment of Erwinia dissolvens and Erwinia nimipressuralis to the genus Enterobacter as Enterobacter dissolvens comb. nov. and Enterobacter nimipressuralis comb. nov. J Clin Microbiol 23:1114–1120. https://doi.org/10.1128/jcm.23.6.1114-1120.1986
Ownsend SM, Hurrell E, Caubilla-Barron J, LocCarrillo C, Forsythe SJ (2008) Characterization of 967 an extended-spectrum beta-lactamase Enterobacter hormaechei nosocomial outbreak, and other Enterobacter hormaechei misidentified as Cronobacter (Enterobacter) sakazakii. Microbiology 154:3659–3667. https://doi.org/10.1099/mic.0.2008/021980-0
Edwards JR, Hayashi JA (1965) Structure of a rhamnolipid from Pseudomonas aeruginosa. Arch Biochem Biophys 111:415–421. https://doi.org/10.1016/0003-9861(65)90204-3
Bazire A, Dufour A (2014) The Pseudomonas aeruginosa rhlG and rhlAB genes are inversely regulated and RhlG is not required for rhamnolipid synthesis. BMC Microbiol 14:1–9. https://doi.org/10.1186/1471-2180-14-160
Andreadou E, Pantazaki AA, Daniilidou M, Tsolaki M (2017) Rhamnolipids, microbial virulence factors, in alzheimer’s disease. J Alzheimer’s Dis 59:209–222. https://doi.org/10.3233/JAD-161020
Abdelmawgoud AM, Lépine F, Déziel E (2014) A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem Biol 21:156–164. https://doi.org/10.1016/j.chembiol.2013.11.010
Six DA, Yuan Y, Leeds JA, Meredith TC (2014) Deletion of the β-acetoacetyl synthase FabY in Pseudomonas aeruginosa induces hypoacylation of lipopolysaccharide and increases antimicrobial susceptibility. Antimicrob Agents Chemother 58:153–161. https://doi.org/10.1128/aac.01804-13
Hošková M, Ježdík R, Schreiberová O, Chudoba J, Šir M, Čejková A, Masák J, Jirků V, Řezanka T (2015) Structural and physiochemical characterization of rhamnolipids produced by Acinetobacter calcoaceticus, Enterobacter asburiae and Pseudomonas aeruginosa in single strain and mixed cultures. J Biotechnol 193:45–51. https://doi.org/10.1016/j.jbiotec.2014.11.014
Yuan Y, Sachdeva M, Leeds JA, Meredith TC (2012) Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by the FabY class of β-ketoacyl acyl carrier protein synthases. J Bacteriol 194:5171–5184. https://doi.org/10.1128/jb.00792-12
Kang Y, Zarzycki-Siek J, Walton CB, Norris MH, Hoang TT (2010) Multiple FadD acyl-CoA synthetases contribute to differential fatty acid degradation and virulence in Pseudomonas aeruginosa. PLoS ONE 5:13557. https://doi.org/10.1371/journal.pone.0013557
Zhu K, Rock CO (2008) RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J Bacteriol 190:3147–3154. https://doi.org/10.1128/jb.00080-08
Dobler L, Vilela LF, Almeida RV, Neves BC (2015) Rhamnolipids in perspective: gene regulatory pathways, metabolic engineering, production and technological forecasting. N Biotechnol 33:123–135. https://doi.org/10.1016/j.nbt.2015.09.005
Burger MM, Glaser L, Burton RM (1963) The enzymatic synthesis of a rhamnose containing glycolipid by extracts of Pseudomonas aeruginosa. J Biol Chem 1:2595–2602. https://doi.org/10.1016/S0021-9258(18)67872-X
Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS (2000) Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiol 2146:2803–2814. https://doi.org/10.1099/00221287-146-11-2803
Olvera C, Goldberg JB, Sánchez R, Soberón-Chávez G (1999) The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol Lett 179:85–90. https://doi.org/10.1016/S0378-1097(99)00381-X
Zegans ME, Wozniak D, Griffin E, Toutainkidd CM, Hammond JH, Garfoot A, Lam JS (2012) Pseudomonas aeruginosa exopolysaccharide Psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob Agents Chemother 56:4112–4122. https://doi.org/10.1128/aac.00373-12
Arun SP, Asim KJ (2020) Utilization of waste frying oil for rhamnolipid production by indigenous Pseudomonas aeruginosa: improvement through co-substrate optimization. J Environ Chem Eng 8:2213–3437. https://doi.org/10.1016/j.jece.2020.104304
Jagruti VJ, Padmini A, Sneha Y, Amit PP, Sandeep BK (2019) Sunflower acid oil-based production of rharmnolipid using Pseudomonas aeruginosa and its application in liquid detergents. J Surfact Deterg 22:463–476. https://doi.org/10.1002/jsde.12255
Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:646–653. https://doi.org/10.1093/nar/gkv1227
Wittgens A, Kovacic F, Müller MM, Gerlitzki M, Santiago-Schübel B, Hofmann D, Tiso T, Blank LM, Henkel M, Hausmann R (2017) Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl Microbiol Biotechnol 101:2865–2878. https://doi.org/10.1007/s00253-016-8041-3
Zhao F, Cui Q, Han S, Dong H, Zhang J, Ma F, Zhang Y (2015) Enhanced rhamnolipid production of Pseudomonas aeruginosa SG by increasing copy number of rhlAB genes with modified promoter. RSC Adv 5:70546–70552. https://doi.org/10.1039/C5RA13415C
Tang T, Fu LH, Xie WH, Luo YZ, Zhang YT, Zhang JZ, Si T (2023) RhlA exhibits dual thioesterase and acyltransferase activities during rhamnolipid biosynthesis. ACS catal 13:5759–5766. https://doi.org/10.1021/acscatal.3c00046
Ochsner UA, Fiechter A, Reiser J (1994) Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 269:19787–19795. https://doi.org/10.1016/S0021-9258(17)32089-6
Tremblay J, Richardson AP, Lépine F, Déziel E (2007) Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 10:2622–2630. https://doi.org/10.1111/j.1462-2920.2007.01396.x
Han L, Liu P, Peng Y, Lin J, Wang Q, Ma Y (2014) Engineering the biosynthesis of novel rhamnolipids in Escherichia coli for enhanced oil recovery. J Appl Microbiol 117:139–150. https://doi.org/10.1111/jam.12515
Ratridewi I, Dzulkarnain SA, Wijaya AB, Barlianto W, Santoso W, Santosaningsih D (2020) Piper betle leaf extract exhibits anti-virulence properties by downregulating rhamnolipid gene expression (rhlC) of Pseudomonas aeruginosa. Open Access Maced J Medical Sci 8:928–931. https://doi.org/10.3889/oamjms.2020.5247
Boles BR, Thoendel M, Singh PK (2005) Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57:1210–1223. https://doi.org/10.1111/j.1365-2958.2005.04743.x
Merced G, Hwan CM, Tian B, Ju X, Kook RJ, Ok KM, You-Hee C, Chul YS, Kaufmann GF (2013) Simultaneous inhibition of rhamnolipid and polyhydroxyalkanoic acid synthesis and biofilm formation in Pseudomonas aeruginosa by 2-bromoalkanoic acids: effect of inhibitor alkyl-chain-length. PLoS One 8:73986. https://doi.org/10.1371/journal.pone.0073986
Reis RS, Pereira AG, Neves BC, Freire DM (2011) Gene regulation of rhamnolipid production in Pseudomonas aeruginosa–a review. Bioresour Technol 102:6377–6384. https://doi.org/10.1016/j.biortech.2011.03.074
Müller MM, Hausmann R (2011) Regulatory and metabolic network of rhamnolipid biosynthesis: traditional and advanced engineering towards biotechnological production. Appl Microbiol Biotechnol 91:251–264. https://doi.org/10.1007/s00253-011-3368-2
Lovaglio RB, Silva VL, Ferreira H, Hausmann R, Contiero J (2015) Rhamnolipids know-how: looking for strategies for its industrial dissemination. Biotechnol Adv 33:1715–1726. https://doi.org/10.1016/j.biotechadv.2015.09.002
Soukarieh F, Williams P, Stocks MJ, Camara M (2018) Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: current position and future perspectives. J Med Chem 61:10385–10402. https://doi.org/10.1021/acs.jmedchem.8b00540
Dekimpe V, Déziel E (2009) Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiol 155:712–723. https://doi.org/10.1099/mic.0.022764-0
Gerardo M, Katy J, Brenda V, Gloria S (2003) Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J Bacteriol 185:5976–5983. https://doi.org/10.1128/jb.185.20.5976-5983.2003
Medina G, Juárez K, Soberón-Chávez G (2003) The Pseudomonas aeruginosa rhlAB operon is not expressed during the logarithmic phase of growth even in the presence of its activator RhlR and the autoinducer N-butyryl-homoserine lactone. J Bacteriol 185:377–380. https://doi.org/10.1128/jb.185.1.377-380.2003
Rampioni G, Falcone M, Heeb S, Frangipani E, Fletcher MP, Dubern JF, Visca P, Leoni L, Cámara M, Williams P (2016) Unravelling the genome-wide contributions of specific 2-alkyl-4-quinolones and PqsE to quorum sensing in Pseudomonas aeruginosa. PLos Pathog 12:e1006029. https://doi.org/10.1371/journal.ppat.1006029
Xiao G, Déziel E, He J, Lépine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG (2006) MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 262:1689–1699. https://doi.org/10.1111/j.1365-2958.2006.05462.x
Ilangovan A, Fletcher M, Rampioni G, Pustelny C, Rumbaugh K, Heeb S, Cámara M, Truman A, Chhabra SR, Emsley J, Williams P (2013) Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR). PLos Pathog. 9:e1003508. https://doi.org/10.1371/journal.ppat.1003508
Chen JW, Lu YJ, Ye F, Zhang HF, Zhou YL, Li JT, Wu Q, Xu XW, Wu QH, Wei B, Zhang HW, Wang H (2022) A small-molecule inhibitor of the anthranilyl-CoA synthetase pqsA for the treatment of multidrug-resistant Pseudomonas aeruginosa. Microbiol Spectr 10:2721–2764. https://doi.org/10.1128/spectrum.02764-21
Farrow JM, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC (2008) PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol 190:7043–7051. https://doi.org/10.1128/jb.00753-08
Shen Y, Vanessa J, Janine S, Ingo F, Stefan W, Erik S, Susanne HU, Wulf B (2009) Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein. Biochem 48:10298–10307. https://doi.org/10.1021/bi900123j
Rampioni G, Pustelny C, Fletcher MP, Wright VJ, Bruce M, Rumbaugh KP, Heeb S, Cámara M, Williams P (2010) Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ Microbiol 12:1659–1673. https://doi.org/10.1111/j.1462-2920.2010.02214.x
Hazan R, He J, Xiao G, Dekimpe V, Apidianakis Y, Lesic B, Astrakas C, Déziel E, Lépine F, Rahme LG (2010) Homeostatic interplay between bacterial cell-cell signaling and iron in virulence. Plos Pathog 6:e1000810. https://doi.org/10.1371/journal.ppat.1000810
Simanek KA, Taylor IR, Richael EK, Lasek-Nesselquist E, Bassler BL, Paczkowski JE (2022) The PqsE-RhlR interaction regulates RhlR DNA binding to control virulence factor production. Microbiol Spectr 10:2108–2121. https://doi.org/10.1128/spectrum.02108-21
Taylor IR, Paczkowski JE, Jeffrey PD, Henke BR, Smith CD, Bassler BL (2021) Inhibitor mimetic mutations in the PqsE enzyme reveal a protein-protein interaction with the quorum-sensing receptor RhlR that is vital for virulence factor production. ACS Chem Biol 16:740–752. https://doi.org/10.1021/acschembio.1c00049
Dandekar A, Greenberg E (2013) Plan B for quorum sensing. Nat Chem Biol 9:292–293. https://doi.org/10.1038/nchembio.1233
Biosurfactants: production and applications (2013) https://www.semanticscholar.org/paper/Chapter-2-Biosurfactants-%3A-Production-and-Reis-Pacheco/fe3c10992d837198ebd394e7a54742cc0c48111e. Accessed 2023
Cai Z, Liu Y, Chen Y, Yam JKH, Chew SC, Chua SL, Wang K, Givskov M, Yang L (2015) RpoN regulates virulence factors of Pseudomonas aeruginosa via modulating the PqsR quorum sensing regulator. Int J Mol Sci 16:28311–28319. https://doi.org/10.3390/ijms161226103
Schulz S, Eckweiler D, Bielecka A, Nicolai T, Franke R, Dotsch A, Hornischer K, Bruchmann S, Duvel J, Haussler S (2015) Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. Plos Pathog 11:e1004744. https://doi.org/10.1371/journal.ppat.1004744
Schuster M, Hawkins AC, Harwood CS, Greenberg E (2004) The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51:973–985. https://doi.org/10.1046/j.1365-2958.2003.03886.x
Chiang SM, Schellhorn HE (2010) Evolution of the RpoS regulon: origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J Mol Evol 70:557–571. https://doi.org/10.1007/s00239-010-9352-0
Liang H, Deng X, Ji Q, Sun F, Shen T, He C (2012) The Pseudomonas aeruginosa global regulator VqsR directly inhibits QscR to control quorum-sensing and virulence gene expression. J Bacterial 194:3098–3108. https://doi.org/10.1128/jb.06679-11
Yang N, Ding S, Chen F, Zhang X, Xia Y, Di H, Cao Q, Deng X, Wu M, Wong CCL (2015) The Crc protein participates in down-regulation of the Lon gene to promote rhamnolipid production and rhl quorum sensing in Pseudomonas aeruginosa. Mol Microbiol 96:526–547. https://doi.org/10.1111/mmi.12954
Zhang L, Gao Q, Chen W, Qin H, Heng ZW, Chen Y, Yang L, Zhang G (2013) Regulation of pqs quorum sensing via catabolite repression control in Pseudomonas aeruginosa. Microbiol 159:1931–1936. https://doi.org/10.1099/mic.0.066266-0
Wenner N, Maes A, Cotado-Sampayo M, Lapouge K (2014) NrsZ: a novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ Microbiol 16:1053–1068. https://doi.org/10.1111/1462-2920.12272
Okkotsu Y, Tieku P, Fitzsimmons LF, Churchill ME, Schurr MJ (2013) Pseudomonas aeruginosa AlgR phosphorylation modulates rhamnolipid production and motility. J Bacteriol 195:5499–5515. https://doi.org/10.1128/jb.00726-13
Kong W, Zhao J, Kang H, Zhu M, Zhou T, Deng X, Liang H (2015) ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res 43:8268–8282. https://doi.org/10.1093/nar/gkv747
Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L (2007) RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol Microbiol 66:1557–1565. https://doi.org/10.1111/j.1365-2958.2007.06029.x
Kang H, Gan J, Zhao J, Kong W, Jing Z, Miao Z, Fan L, Song Y, Jin Q, Liang H (2016) Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing. Nucleic Acids Res 45:699–710. https://doi.org/10.1093/nar/gkw954
Ren B, Shen H, Lu ZJ, Liu H, Xu Y (2014) The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLos One 9:e89653. https://doi.org/10.1371/journal.pone.0089653
Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Cámara M, Williams P, Haas D (2004) Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol 186:2936–2945. https://doi.org/10.1128/jb.186.10.2936-2945.2004
Cao L, Wang Q, Zhang J, Li C, Yan X, Lou X, Xia Y, Hong Q, Li S (2012) Construction of a stable genetically engineered rhamnolipid-producing microorganism for remediation of pyrene-contaminated soil. W J Micro Bio 28:2783–2790. https://doi.org/10.1007/s11274-012-1088-0
Kahraman H, Erenler S (2012) Rhamnolipid production by Pseudomonas aeruginosa engineered with the Vitreoscilla hemoglobin gene. Appl Biochem Micro 48:188–193. https://doi.org/10.1134/S000368381202007X
Zhao F, Shi R, Zhao J, Li G, Bai X, Han S, Zhang Y (2015) Heterologous production of Pseudomonas aeruginosa rhamnolipid under anaerobic conditions for microbial enhanced oil recovery. J App Microbiol 118:379–389. https://doi.org/10.1111/jam.12698
Cha M, Lee N, Kim M, Kim M, Lee S (2008) Heterologous production of Pseudomonas aeruginosa EMS1 biosurfactant in Pseudomonas putida. Bioresour Technol 99:2192–2199. https://doi.org/10.1016/j.biortech.2007.05.035
Cabrera-Valladares N, Richardson AP, Olvera C, Treviño LG, Déziel E, Lépine F, Soberón-Chávez G (2006) Monorhamnolipids and 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Appl Microbiol Biotechnol 73:187–194. https://doi.org/10.1007/s00253-006-0468-5
Wang Q, Fang X, Bai B, Liang X, Shuler PJ, Goddard WA, Tang Y (2007) Engineering bacteria for production of rhamnolipid as an agent for enhanced oil recovery. Biotechnol Bioeng 98:842–853. https://doi.org/10.1002/bit.21462
Ochsner UA, Reiser J, Fiechter A, Witholt B (1995) Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl Environ Microbiol 6:3503–3506. https://doi.org/10.1128/aem.61.9.3503-3506.1995
Solaiman DK, Ashby RD, Gunther NW IV, Zerkowski JA (2015) Dirhamnose-lipid production by recombinant nonpathogenic bacterium Pseudomonas chlororaphis. Appl Environ Microbiol 99:4333–4342. https://doi.org/10.1007/s00253-015-6433-4
Gong ZJ, Peng YF, Zhang YT, Song GT, Chen WJ, Jia S, Wang QH (2015) Construction and optimization of Escherichia coli for producing rhamnolipid biosurfactant. Chin J Biotechnol 31:1050–1062. https://doi.org/10.4014/jmb.1104.04048
Setoodeh P, Jahanmiri A, Eslamloueyan R, Niazi A, Ayatollahi SS, Aram F, Mahmoodi M, Hortamani A (2014) Statistical screening of medium components for recombinant production of Pseudomonas aeruginosa ATCC 9027 rhamnolipids by nonpathogenic cell factory Pseudomonas putida KT2440. Mol Biotechnol 56:175–191. https://doi.org/10.1007/s12033-013-9693-1
Wittgens A, Tiso T, Arndt TT, Wenk P, Hemmerich J, Müller C, Wichmann R, Küpper B, Zwick M, Wilhelm S, Hausmann R, Syldatk C, Rosenau F, Blank LM (2011) Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Fact 10:80. https://doi.org/10.1186/1475-2859-10-80
Tavares LF, Silva PM, Junqueira M, Mariano D, Nogueira FC, Domont GB, Freire DM, Neves BC (2013) Characterization of rhamnolipids produced by wild-type and engineered Burkholderia kururiensis. Appl Microbiol Biotechnol 97:1909–1921. https://doi.org/10.1007/s00253-012-4454-9
Funding
The project was supported by Natural Foundation of Zhejiang Province (LGF21H300003), Key Laboratory of Tropical Marine Ecosystem and Bioresource, MNR (2021QN03), National Natural Science Foundation of China (No. 42276137).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare that they have no competing interests.
Ethical Approval
No experiment was conducted on animals. Since, the current work is entirely based upon the previous data and sample, this study does not warrant any ethical approval certificate.
Consent to Participate
Not applicable for this work.
Consent to Publish
Not applicable for this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., Yu, X., Lu, X. et al. Biosynthesis and Gene Regulation of Rhamnolipid Congeners. Curr Microbiol 80, 302 (2023). https://doi.org/10.1007/s00284-023-03405-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03405-x