Abstract
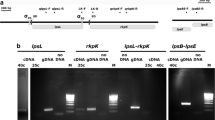
Lysobacter capsici X2-3, a plant growth-promoting rhizobacteria (PGPR), was isolated from wheat rhizosphere and has inhibitory effects against a wide range of pathogens. One important characteristic of L. capsici is its ability to produce diverse antibiotics and lytic enzymes. The GntR family of transcription factors is a common transcription factor superfamily in bacteria that has fundamental roles in bacterial metabolism regulation. However, the GntR family transcription factor in Lysobacter has not been identified. In this study, to obtain an understanding of the GntR/HutC gene function in L. capsici X2-3, a random Tn5-insertion mutant library of X2-3 was constructed to select genes showing pleiotropic effects on phenotype. We identified a Tn5 mutant with an insertion in LC4356 that showed reduced biofilm levels, and sequence analysis indicated that the inserted gene encodes a GntR/HutC family transcription regulator. Furthermore, the LC4356 mutant showed reduced extracellular polysaccharide (EPS) production, diminished twitching motility and decreased survival under UV radiation and high-temperature. The RT‒qPCR results indicated that the pentose phosphate pathway-related genes G6PDH, 6PGL and PGDH were upregulated in the LC4356 mutant. Thus, since L. capsici is an efficient biocontrol agent for crop protection, our findings provide fundamental insights into GntR/HutC and will be worthwhile to improve PGPR biocontrol efficacy.





Similar content being viewed by others
Data Availability
If reasonable request is made, the corresponding author will provide the datasets used or analyzed during the study.
Code Availability
Not applicable.
References
Afoshin AS, Kudryakova IV, Borovikova AO, Suzina NE, Toropygin IY, Shishkova NA et al (2020) Lytic potential of Lysobacter capsici VKM B-2533(T): bacteriolytic enzymes and outer membrane vesicles. Sci Rep 10:9944. https://doi.org/10.1038/s41598-020-67122-2
Fang BZ, Xie YG, Zhou XK, Zhang XT, Liu L, Jiao JY et al (2020) Lysobacter prati sp. nov., isolated from a plateau meadow sample. Antonie Van Leeuwenhoek 113:763–772. https://doi.org/10.1007/s10482-020-01386-6
Yu L, Su W, Fey PD, Liu F, Du L (2018) Yield improvement of the anti-MRSA antibiotics WAP-8294A by CRISPR/dCas9 combined with refactoring self-protection genes in Lysobacter enzymogenes OH11. ACS Synth Biol 7:258–266. https://doi.org/10.1021/acssynbio.7b00293
Park JH, Kim R, Aslam Z, Jeon CO, Chung YR (2008) Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int J Syst Evol Microbiol 58:387–392. https://doi.org/10.1099/ijs.0.65290-0
Zhao Y, Qian G, Ye Y, Wright S, Chen H, Shen Y et al (2016) Heterocyclic aromatic N-Oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org Lett 18:2495–2498. https://doi.org/10.1021/acs.orglett.6b01089
Puopolo G, Raio A, Zoina A (2010) Identification and characterization of Lysobacter capsici strain PG4: a new plant health-promoting rhizobacterium. J Plant Pathol 92:157–164. https://doi.org/10.4454/jpp.v92i1.25
Puopolo G, Cimmino A, Palmieri MC, Giovannini O, Evidente A, Pertot I (2014) Lysobacter capsici AZ78 produces cyclo(L-Pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J Appl Microbiol 117:1168–1180. https://doi.org/10.1111/jam.12611
Brescia F, Marchetti-Deschmann M, Musetti R, Perazzolli M, Pertot I, Puopolo G (2020) The rhizosphere signature on the cell motility, biofilm formation and secondary metabolite production of a plant-associated Lysobacter strain. Microbiol Res 234:126424. https://doi.org/10.1016/j.micres.2020.126424
Arya G, Pal M, Sharma M, Singh B, Singh S, Agrawal V et al (2021) Molecular insights into effector binding by DgoR, a GntR/FadR family transcriptional repressor of D-galactonate metabolism in Escherichia coli. Mol Microbiol 115(4):591–609. https://doi.org/10.1111/mmi.14625
Li Z, Xiang Z, Zeng J, Li Y, Li J (2018) A GntR family transcription factor in Streptococcus mutans regulates biofilm formation and expression of multiple sugar transporter genes. Front Microbiol 9:3224. https://doi.org/10.3389/fmicb.2018.03224
Vindal V, Suma K, Ranjan A (2007) GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC Genom 8:289. https://doi.org/10.1186/1471-2164-8-289
Zhou X, Yan Q, Wang N (2017) Deciphering the regulon of a GntR family regulator via transcriptome and ChIP-exo analyses and its contribution to virulence in Xanthomonas citri. Mol Plant Pathol 18:249–262. https://doi.org/10.1111/mpp.12397
Abeywickrama TD, Perera IC (2021) In silico characterization and virtual screening of GntR/HutC family transcriptional regulator MoyR: a potential monooxygenase regulator in Mycobacterium tuberculosis. Biology 10:1241. https://doi.org/10.3390/biology10121241
Aravind L, Anantharaman V (2003) HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold. FEMS Microbiol Lett 222:17–23. https://doi.org/10.1016/s0378-1097(03)00242-8
Naren N, Zhang XX (2020) Global regulatory roles of the histidine-responsive transcriptional repressor HutC in Pseudomonas fluorescens SBW25. J Bacteriol 202:e00792-e819. https://doi.org/10.1128/jb.00792-19
Rietsch A, Wolfgang MC, Mekalanos JJ (2004) Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect Immun 72:1383–1390. https://doi.org/10.1128/iai.72.3.1383-1390.2004
Cao ZL, Tan TT, Zhang YL, Han L, Hou XY, Ma HY et al (2018) NagR(Bt) is a pleiotropic and dual transcriptional regulator in Bacillus thuringiensis. Front Microbiol 9:1899. https://doi.org/10.3389/fmicb.2018.01899
Liao CH, Xu Y, Rigali S, Ye BC (2015) DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea. Appl Microbiol Biotechnol 99:10215–10224. https://doi.org/10.1007/s00253-015-6892-7
Yi JL, Wang J, Li Q, Liu ZX, Zhang L, Liu AX et al (2015) Draft genome sequence of the bacterium Lysobacter capsici X2–3, with a broad spectrum of antimicrobial activity against multiple plant-pathogenic microbes. Genome Announc 3:e00589-e615. https://doi.org/10.1128/genomeA.00589-15
Zhao D, Wang H, Li Z, Han S, Han C, Liu A (2022) LC_glucose-inhibited division protein is required for motility, biofilm formation, and stress response in Lysobacter capsici X2–3. Front Microbiol 13:840792. https://doi.org/10.3389/fmicb.2022.840792
Obranić S, Babić F, Maravić-Vlahoviček G (2013) Improvement of pBBR1MCS plasmids, a very useful series of broad-host-range cloning vectors. Plasmid 70:263–267. https://doi.org/10.1016/j.plasmid.2013.04.001
Rehl JM, Shippy DC, Eakley NM, Brevik MD, Sand JM, Cook ME et al (2013) GidA expression in Salmonella is modulated under certain environmental conditions. Curr Microbiol 67:279–285. https://doi.org/10.1007/s00284-013-0361-2
Zhang Y, Gao J, Wang L, Liu S, Bai Z, Zhuang X et al (2018) Environmental adaptability and quorum sensing: iron uptake regulation during biofilm formation by Paracoccus denitrificans. Appl Environ Microbiol 84:00865–00918. https://doi.org/10.1128/aem.00865-18
Javvadi S, Pandey SS, Mishra A, Pradhan BB, Chatterjee S (2018) Bacterial cyclic β-(1,2)-glucans sequester iron to protect against iron-induced toxicity. EMBO Rep 19:172–186. https://doi.org/10.15252/embr.201744650
Hosseinidoust Z, Tufenkji N, van de Ven TG (2013) Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl Environ Microbiol 79:2862–2871. https://doi.org/10.1128/AEM.03817-12
Dye KJ, Vogelaar NJ, O’Hara M, Sobrado P, Santos W, Carlier PR et al (2022) Discovery of two inhibitors of the type IV pilus assembly ATPase PilB as potential antivirulence compounds. Microbiol Spectr 10:e0387722. https://doi.org/10.1128/spectrum.03877-22
Li D, Shibata Y, Takeshita T, Yamashita Y (2014) A novel gene involved in the survival of Streptococcus mutans under stress conditions. Appl Environ Microbiol 80:97–103. https://doi.org/10.1128/aem.02549-13
Su HZ, Wu L, Qi YH, Liu GF, Lu GT, Tang JL (2016) Characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Sci Rep 6:19862. https://doi.org/10.1038/srep19862
Li J, Wang N (2011) The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol Plant Pathol 12:381–396. https://doi.org/10.1111/j.1364-3703.2010.00681.x
Antar A, Lee MA, Yoo Y, Cho MH, Lee SW (2020) PXO_RS20535, encoding a novel response regulator, is required for chemotactic motility, biofilm formation, and tolerance to oxidative stress in Xanthomonas oryzae pv. oryzae. Pathogens 9:956. https://doi.org/10.3390/pathogens9110956
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Tomada S, Puopolo G, Perazzolli M, Musetti R, Loi N, Pertot I (2016) Pea broth enhances the biocontrol efficacy of Lysobacter capsici AZ78 by triggering cell motility associated with biogenesis of type IV pilus. Front Microbiol 7:1136. https://doi.org/10.3389/fmicb.2016.01136
Pawlik KJ, Zelkowski M, Biernacki M, Litwinska K, Jaworski P, Kotowska M (2021) GntR-like SCO3932 protein provides a link between actinomycete integrative and conjugative elements and secondary metabolism. Int J Mol Sci 22:11867. https://doi.org/10.3390/ijms222111867
Daddaoua A, Corral-Lugo A, Ramos JL, Krell T (2017) Identification of GntR as regulator of the glucose metabolism in Pseudomonas aeruginosa. Environ Microbiol 19:3721–3733. https://doi.org/10.1111/1462-2920.13871
Mielich-Süss B, Lopez D (2015) Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17:555–565. https://doi.org/10.1111/1462-2920.12527
Zhou T, Nan B (2017) Exopolysaccharides promote Myxococcus xanthus social motility by inhibiting cellular reversals. Mol Microbiol 103:729–743. https://doi.org/10.1111/mmi.13585
Martín-Rodríguez AJ, Villion K, Yilmaz-Turan S, Vilaplana F, Sjöling A, Römling U (2021) Regulation of colony morphology and biofilm formation in Shewanella algae. Microb Biotechnol 14:1183–1200. https://doi.org/10.1111/1751-7915.13788
Limoli DH, Jones CJ, Wozniak DJ (2015) Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.MB-0011-2014
Palmer SR, Ren Z, Hwang G, Liu Y, Combs A, Söderström B et al (2019) Streptococcus mutans yidC1 and yidC2 impact cell envelope biogenesis, the biofilm matrix, and biofilm biophysical properties. J Bacteriol 201:e00396-e418. https://doi.org/10.1128/jb.00396-18
Sorroche F, Bogino P, Russo DM, Zorreguieta A, Nievas F, Morales GM et al (2018) Cell Autoaggregation, biofilm formation, and plant attachment in a Sinorhizobium meliloti lpsB mutant. Mol Plant Microbe Interact 31:1075–1082. https://doi.org/10.1094/mpmi-01-18-0004-r
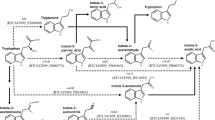
Bradfield MF, Nicol W (2016) The pentose phosphate pathway leads to enhanced succinic acid flux in biofilms of wild-type Actinobacillus succinogenes. Appl Microbiol Biotechnol 100:9641–9652. https://doi.org/10.1007/s00253-016-7763-6
Li C, Hurley A, Hu W, Warrick JW, Lozano GL, Ayuso JM et al (2021) Social motility of biofilm-like microcolonies in a gliding bacterium. Nat Commun 12:5700. https://doi.org/10.1038/s41467-021-25408-7
Patra P, Kissoon K, Cornejo I, Kaplan HB, Igoshin OA (2016) Colony expansion of socially motile Myxococcus xanthus cells is driven by growth, motility, and exopolysaccharide production. PLoS Comput Biol 12:e1005010. https://doi.org/10.1371/journal.pcbi.1005010
Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G et al (2014) Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl Environ Microbiol 80:6724–6732. https://doi.org/10.1128/aem.01237-14
Gao T, Ding M, Yang CH, Fan H, Chai Y, Li Y (2019) The phosphotransferase system gene ptsH plays an important role in MnSOD production, biofilm formation, swarming motility, and root colonization in Bacillus cereus 905. Res Microbiol 170:86–96. https://doi.org/10.1016/j.resmic.2018.10.002
Masmoudi F, Abdelmalek N, Tounsi S, Dunlap CA, Trigui M (2019) Abiotic stress resistance, plant growth promotion and antifungal potential of halotolerant bacteria from a Tunisian solar saltern. Microbiol Res 229:126331. https://doi.org/10.1016/j.micres.2019.126331
Morcillo RJL, Manzanera M (2021) The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 11:337. https://doi.org/10.3390/metabo11060337
Pérez-Burgos M, Søgaard-Andersen L (2020) Biosynthesis and function of cell-surface polysaccharides in the social bacterium Myxococcus xanthus. Biol Chem 401:1375–1387. https://doi.org/10.1515/hsz-2020-0217
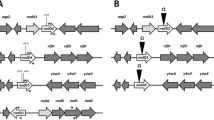
Liu GF, Wang XX, Su HZ, Lu GT (2021) Progress on the GntR family transcription regulators in bacteria. Yi Chuan 43:66–73. https://doi.org/10.16288/j.yczz.20-245
Abdalla AK, Ayyash MM, Olaimat AN, Osaili TM, Al-Nabulsi AA, Shah NP et al (2021) Exopolysaccharides as antimicrobial agents: mechanism and spectrum of activity. Front Microbiol 12:664395. https://doi.org/10.3389/fmicb.2021.664395
Funding
This work was supported by Outstanding Youth Foundation of Shandong Province [Grant Number ZR2021YQ20], the Shandong Modern Agricultural Industry Technology System [Grant Number SDAIT-04-08], Key Research and Development Program of Shandong Province [Grant Number 2019JZZY020608], Weifang Tobacco Company Science and Technology Project [Grant Number 2022-36].
Author information
Authors and Affiliations
Contributions
The study was conceived by JW, the majority of the experiments were conducted by JW and DZ, and some were assisted by HW, the data were analyzed by XZ, DZ, CH and AL drafted the manuscript. Before submission, all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
There are no human subjects or animals used in this article.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, D., Wang, J., Wang, H. et al. The Transcription Regulator GntR/HutC Regulates Biofilm Formation, Motility and Stress Tolerance in Lysobacter capsici X2-3. Curr Microbiol 80, 281 (2023). https://doi.org/10.1007/s00284-023-03390-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03390-1