Abstract
Candida parapsilosis is a common cause of candidiasis among hospitalized patients, often surpassing Candida albicans. Due to the recent increase in C. parapsilosis infections, there is an urgent need for rapid, sensitive, and real-time on-site detection of nucleic acids for timely diagnosis of candidiasis. We developed an assay for detection of C. parapsilosis by combining recombinase polymerase amplification (RPA) with a lateral flow strip (LFS). The RPA-LFS assay was used to amplify the beta-1,3-glucan synthase catalytic subunit 2 (FKS2) gene of C. parapsilosis with a primer–probe set optimized by introducing base mismatches (four bases modified by the probe and one by the reverse primer) to achieve specific and sensitive detection of clinical samples. The RPA assays can rapidly amplify and visualize a target gene within 30 min, while the entire process can be completed within 40 min by pre-processing the sample. The product of RPA has two chemical labels, FITC and Biotin, of the amplification product can be carefully on the strip. The sensitivity and specificity of the RPA-LFS assay were determined by analysis of 35 common clinical pathogens and 281 clinical samples against quantitative PCR. The results confirmed that the proposed RPA-LFS assay is a reliable molecular diagnostic method for the detection of C. parapsilosis to meet the urgent need for rapid, specific, sensitive, and portable field testing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Candida parapsilosis is a common cause of candidiasis in low birth weight neonates and critically ill patients who are mainly infected during invasive procedures, such as central venous catheter placement, surgery, and parenteral nutrition [1, 2]. The incidence of invasive candidiasis has increased in recent decades and the detection rate of candidiasis caused by C. parapsilosis exceeds that of Candida albicans in some hospitals in Europe, Asia, and South America [3]. Incidences of candidemia increased from 1.97 episodes/10,000 patient-days in 2008 to 4.59/10,000 patient-days in 2016, 30 days mortality was 32.4% [4].
Traditional fungal culture methods are the gold standard for the diagnosis of fungal infectious diseases. The continued development of molecular diagnostic techniques has been applied in the clinical detection of C. parapsilosis [5]. Among these techniques include polymerase chain reaction (PCR)-restriction fragment length polymorphism based on DNA amplification [6], random amplified polymorphic DNA [7], real-time PCR [8], PCR for intron length analysis of polymorphisms [9], matrix-assisted laser desorption ionization time-of-flight mass spectrometry [10], and sequencing analysis of pan-fungal markers. Meanwhile, isothermal amplification is more frequently utilized in field testing, as this method is not limited by instrumentation and laboratory settings. The main isothermal amplification techniques currently available include loop-mediated isothermal amplification (LAMP) [11], nuclear acid sequence-based amplification (NASBA) [12], rolling circle amplification (RCA) [13], single primer isothermal amplification [14] (SPIA), helicase-dependent isothermal DNA amplification (HDA) [15], and strand displacement amplification (SDA) [16]. Recombinase polymerase amplification (RPA) is a recently developed technique that merges the advantages of thermostatic amplification and compensating for the shortcomings as a rapid, specific, sensitive, and portable diagnostic assay.
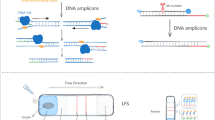
RPA uses recombinase junctions that bind specific primers to open the double-stranded helix to bind to the target fragment, single-stranded DNA binding protein wraps single-stranded DNA with high affinity to protect against degradation and secondary structure formation, and the polymerase of Bacillus subtilis recognizes the 3′ end of the primer for rapid amplification (Supplementary Fig. S1A and B), which can produce a large number of amplification products in about 20 min at 30–45 °C [17]. The amplification products of RPA can be detected by gel electrophoresis, a fluorescence detector, or with the use of a lateral flow strip (LFS). However, gel electrophoresis and fluorescence detection is limited to laboratory settings. In contrast, LFS is suitable for in situ detection in field conditions and the results can be directly observed without a dedicated display device or the need for sophisticated thermal cycling equipment and highly trained technicians. Visualization of a LFS is dependent on labeling of the 5′ and 3′ ends of the probe with fluorescein isothiocyanate (FITC) and C3 blocking sites, respectively, while the reverse primer is labeled with biotin. Binding of the probe to the amplification strand is performed by cleavage of the middle tetrahydrofuran (THF) site of the probe by endonuclease IV (nfo gene of Escherichia coli K-12) to release the 3′ end of the probe for extension and the 5′ and 3′ ends of the final amplification product are labeled with FITC and biotin, respectively. Gold nanoparticle (AuNP)-labeled anti-FITC antibody binds to the amplification product and is subsequently captured and aggregated by streptavidin on the detection line to produce a red positive signal, while the secondary antibody on the quality control line captures the AuNP-labeled anti-FITC antibody without binding the amplification product and produces a red signal regardless of the presence or absence of the amplification product (Supplementary Fig. S1C) [18, 19]. The RPA-LFS assay can achieve fast response times and good accuracy for the diagnosis of various infectious diseases.
In our study, RPA and qPCR primers were designed for the diagnosis of C. parapsilosis by FKS2 sequence. To test the effectiveness of the assay system, primer and probe specificity screening, sensitivity testing, specificity analysis, and testing of clinical samples was performed to ensure that our established method could be used for the diagnosis of Candida infections caused by C. parapsilosis.
Methods
Strain acquisition
Candida parapsilosis (C. parapsilosis means C. parapsilosis sensu stricto unless otherwise specified) ATCC 22019/90018/200954/7330 was purchased from Shanghai Covey Chemical Technology Co., Ltd. (Shanghai, China) and 15 strains of C. parapsilosis were isolated from clinical samples collected from 2020 to 2021, and the authenticity of the strain was verified by qPCR. The specificity of the RPA-LFS assay was verified based on the FKS2 gene (GenBank: EU221326.1) of 35 common pathogens stored in our laboratory, which included Candida tropicalis ATCC 20962, C. albicans ATCC 10231, Candida auris, Candida dubliniensis, Candida krusei, Candida glabrata, Aspergillus fumigatus, Cryptococcus neoformans ATCC 14116, Enterococcus faecium, E. coli O157, Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus capitis, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus saprophyticus, Staphylococcus warneri, Stenotrophomonas maltophilia, Streptococcus pneumonia, Viridans streptococci, Klebsiella pneumoniae, and Acinetobacter baumannii ATCC 1960, Candida metapsilosis, Candida orthopsilosis, Cryptococcus gattii, Acinetobacter calcoaceticus, Acinetobacter lwoffi, Acinetobacter haemolytius, Acinetobacter junii, Acinetobacter johnsonii, Enterobacter cloacae, Mycobacterium tuberculosis H37Ra, Listeria monocytogenes, Neisseria meningitidis. In addition to isolating 15 strains of C. parapsilosis and 35 common pathogens stored in our laboratory, an additional 281 strains in total (Blood specimen 124, sputum specimen 107, pus specimen 28, dermal specimen 22) were collected from patients with suspected Candida infection.
Genomic DNA extraction
All bacterial strains were boiled at 100 °C for 10 min to release DNA for use as templates and the concentration was checked by nucleic acid quantification (UV260/280 ratio). If not otherwise specified, 1 μl of 107 copies/μl of heat-treated culture was used as a template. For C. parapsilosis and other fungi, genomic DNA was extracted and purified from cultures or clinical samples using the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions and quantified using a Invitrogen™ Qubit™ 4 Fluorometer (Thermo Fisher Scientific).
Object and Probe Design and Screening
Two primer pairs based on the FKS2 gene were designed for RPA using Primer Premier 5 software (Premier Biosoft, Palo Alto, CA, USA) with the following parameters: product size, 80–150 bp; primer size, 30–35 bp; complementary pairing, ≤ 3 consecutive bases at the 3ʹ end; maximum hairpin fraction, 5; maximum primer-dimer fraction, 5; and maximum poly-X, 5. The primers were designed based on sequences retrieved from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). The species specificity of the primers and probes were confirmed using the NCBI primer designing tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast). The performance of the forward primer, which was extended backward by 15–23 bp, was evaluated to theoretically avoid the formation of dimers and hairpin structures using Primer Premier 5 software. The size of the probe was 46–53 bp with a GC content of 30–80% and melting temperature of 57–80 °C. The 5′ end of the probe was labeled with FITC, the 3′ end was closed with a C3 spacer, the bases in the middle of the probe (at least 30 bases before the THF site and at least 15 afterward) were replaced with THF, and the 5′ end of the reverse primer was labeled with biotin.
RPA Reaction
RPA reactions were performed using the TwistAmp® Liquid DNA Amplification Kit (TwistDx Inc., Maidenhead, UK) in accordance with the manufacturer’s instructions. Each 50-µl reaction system contained 25 µl of 2 × reaction buffer, 5 µl of 10 × E-mix, 2.5 µl of 20/ core mix, 2.4 µl of 10 µM forward primer, 2.4 µl of 10 µM reverse primer, and 9.2 µl of distilled water, in addition to 2.5 μl of 280 mM magnesium acetate and 1 μl of template to the top of the reaction tube. After a short centrifugation step, the reaction mixture was incubated at 37 °C for 30 min. The RPA amplification products were purified using the PCR Cleaning Kit (Shanghai MEIJI Biotechnology Co. Ltd.) and then separated by electrophoresis on a 2% agarose gel.
RPA-LFS Assay
RPA reactions were performed using the Twist Amp® DNA amplification nfo kit (TwistDx Ltd.) in accordance with the manufacturer’s instructions. Each 50-μL reaction mixture contained 2.1 μl of each primer (10 μM), 0.6 μl of the probe (10 μM), 2.0 μl of the template, and other standard reaction components. Primers and probes were synthesized by Anhui General Biotechnology Co., Ltd (Chuzhou, China). Magnesium acetate (280 mM, 2.5 μl) was added to initiate the reaction prior to incubation of the reaction mixture at 37 °C for 20 min. Then, 5 μl of the amplification product were diluted 20-fold and spotted on the LFS (USTAR Biotechnology Co., Ltd., Hangzhou, China). The LFS consisted of a sample pad, a gold-labeled antibody pad (soaked with mouse-derived AuNP-labeled anti-FITC antibody), a test line (coated with streptavidin), a control line (coated with anti-mouse antibody), and an absorption pad, arranged by the solvent migration pathway. The RPA amplification product was added to the sample pad of the LFS and the LFS was submerged into 100 μl of solvent for approximately 2 min until the test and control lines became visible.
Sensitivity and Specificity of the RPA-LFS Assay
A tenfold gradient dilution was tested from 5.0 × 102 to 5.0 × 107 copies/µl (reaction volume of 50 µl containing 1 µl of C. parapsilosis inactivation solution) and 107 copies/µl of an inactivating solution of another common pathogen (C. albicans) were prepared for the RPA-LFS reaction. The lower limit of detection (LOD) of the method was determined with a probit regression analysis of 10 independent experiments. To verify the specificity of the assay system, 35 common clinical fungi and bacterial were selected for RPA-LFS testing.
Quantitative PCR (qPCR) analysis
The primers and probe for qPCR analysis are listed in Table 1. Specific primers and probe were targeted to FKS2 of C. parapsilosis for qPCR detection. Each qPCR reaction mixture consisted of 12.5 μl of MonAmp™ Taqman qPCR mixture (Tiangen Biotechnology Co., Ltd., Beijing, China), 0.5 μM forward and reverse primers, 0.2 μM probe, 1 μl of genomic DNA, and distilled water to a final volume of 25 μl. The cycling program consisted of an initial denaturation step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 55 °C for 60 s. The qPCR reactions were conducted using a LightCycler® 480 System (Roche Diagnostics GmbH, Mannheim, Germany).
Conventional Culture
According to Author Wang et al. [20], the 281 clinical samples were incubated on Stachybotrys medium for 24–48 h and then further inoculated on Candida chromogenic medium. All clinical samples (original specimens) were tested simultaneously using three methods: RPA-LFS, qPCR, and conventional culture.
Results
Primer Screening and Validation
FKS2 was selected from the C. parapsilosis genome as a target gene for RPA-LFS detection. NCBI Primer-BLAST yielded two potential primer pairs for amplification of FKS2 (Supplementary Table 1). These primers were initially screened by amplification of target gene fragments and a no template control (NTC). The amplification products were separated by electrophoresis on an agarose gel to compare the amplification performance of the target gene with the formation of primer-dimers by the NTC. The best amplification performance was obtained with the primer pair F2/R2 and no primer dimer was formed (Fig. 1a). The candidate probe was obtained by extending the 3′ end of the forward primer F2 by 16 bp. All possible cross-dimers generated by this probe and the reverse primer were predicted for subsequent modification of the base sequence (Supplementary Table 1) until no dimer could be formed (Fig. 1b). As shown in the Fig. 1b, there are four potential risks of dimer formation in theory before base replacement. Finally, the five forward primers were screened and tested upstream of the probe. The LFS results showed that the primer/probe sets F3/P/R2B, F4/P/R2B, F5/P/R2B, and F6/P/R2B effectively amplified the target gene fragment, while the negative controls F3/P/R2B, F4/P/R2B, and F5/P/R2B (NTC) produced false-positive signals. Hence, only F6/P/R2B met the requirements of the assay and had effectively amplified the FKS2 fragment without amplification as a NTC (Fig. 1c). Therefore, the primer/probe set F6/P/R2B was used in the subsequent experiments.
Screening of the primer–probe sets. a RPA results for two different primer sets targeting FKS2. The primer set is specified at the top of the corresponding lane. A NTC was used in all reactions, which were performed at 37 °C for 20 min. The results of three independent experiments are shown. b Pairwise analysis and sequence modification of the primer–probe sets for detection of FKS2 using Primer Premier 5 software. The associated DNA base substitutions of the probe and primers are shown. The DNA strands are shown as horizontal lines and the matching bases are indicated by vertical lines. Molecular markers are listed in (b). c Validity of the primer–probe sets for the RPA-LFS assay. The primer set is specified at the top of the corresponding lane. The positions of the test and control lines are shown on the right. All reactions were performed at 37 °C for 20 min. The results of three independent experiments are shown
RPA Reaction Temperature and LFS Chromogenic Time
The temperature parameters were set to 35–45 °C for 30 min. As shown in Fig. 2a, the signal on the test line appears on each temperature gradient; the line is more obvious at 37 °C. Time parameters were set from 5 to 30 min. As shown in Fig. 2b, the pink test line appears at 10 min, and the line becomes clearer from 15 to 30 min. There is no significant difference between the 20 and 30 min lines. Therefore, a response time of 20 min was chosen for the RPA-LFS test of C. parapsilosis. Optimal conditions for C. parapsilosis RPA-LFS were determined to be incubation at 37 °C for 20 min.
Sensitivity of the RPA-LFS Assay
To determine the LOD of C. parapsilosis, a tenfold gradient dilution was tested from 5.0 × 102 to 5.0 × 107 copies/µl. A red band appeared on the test line at 5.0 × 102 copies/µl. Moreover, the red band became darker with an increasing concentration of the C. parapsilosis solution (Fig. 3a). To determine whether the system is resistant to interference from other fungal DNA, 5.0 × 107 copies/µl of C. albicans were added to the RPA reaction. The results showed that the C. albicans inactivation solution did not interfere with the detection of C. parapsilosis (Fig. 3b). Hence, the LOD of the RPA-LFS system was 5.0 × 102 copies/50 µl per reaction and the presence of other fungi did not interfere with the detection sensitivity. However, not all assays yielded positive results when using 5.0 × 102 (12.5%, 1/8 samples) and 5.0 × 103 (75% positive results, 6/8 samples) copies, each samples had been finished ten independent experiments and the results of them were consistent. The results had been listed in Supplementary Table 2. To determine the exact LOD of the RPA-LFS assay, probit regression analysis of data from eight independent assays was performed using SPSS software (SPSS, Inc., Chicago, IL, USA). At 95% probability, the lowest LOD was 5.85 × 103 copies per reaction (Fig. 3c).
Determination of the LOD of the RPA-LFS assay for detection of C. parapsilosis. a The LOD of the C. parapsilosis RPA-LFS assay with the primer–probe set F6/P/R2B was determined from eight independent assays using an inactivating solution of C. parapsilosis with serial dilutions from 5.0 × 102 to 5.0 × 107 copies. The results of the RPA-LFS assay are shown. The number of templates is indicated at the top of the bar graph. b The results of the RPA-LFS assay using primer–probe set F6/P/R2B and 5.0 × 107 copies of C. albicans as interference. c Probit regression analysis of data collected from eight replicates was performed using SPSS software
Interspecies Specificity of the RPA-LFS Assay
To confirm the inclusivity and specificity of the primer–probe set F6/P/R2B, four reference strains, 17 clinical isolates, and other pathogenic bacteria were amplified (Table 1). We tested the nucleic acid concentration of genomic DNA extracted by boiling method and carried out three technical and three biological repeats respectively. The results are shown in Supplementary Table 1. The four reference strains and 17 clinical isolates produced positive results (Fig. 4), while the results for all other pathogenic cultures were negative (Fig. 5a and b), indicating that the primer–probe set was specific for C. parapsilosis without cross-reactivity with other pathogens.
Validation of the specificity of the primer–probe set F6/P/R2B for detection of C. parapsilosis. #1-#17 refers to 17 C. parapsilosis isolates from clinical samples. The positions of the test and control lines are marked on the right side of the bar graph. Reactions were performed at 37 °C for 20 min. The results of three independent experiments are shown. NTC, no template control
Specificity of the primer–probe set F6/P/R2B. a, b C. parapsilosis ATCC 22,019 was used as a positive control against other pathogenic bacteria. Species are identified at the top of each strip. The positions of the test and control lines are marked on the right side of the bars. Reactions were performed at 37 °C for 20 min. The results of three independent experiments are shown. NTC, no template control
Analysis of Clinical Samples
For verification, 281 clinical samples were collected for the RPA-LFS, qPCR, and traditional culture assays, respectively. The detection rate of 89 C. parapsilosis isolates of all the samples 281 clinical was 31.7%, consistent with the detection results of qPCR and traditional culture methods. The experimental results showed that the accuracy of the RPA-LFS assay was the same of that of qPCR and consistent with traditional culture methods.
Discussion
The incidence of hospital-acquired infections caused by Candida spp. continues to increase, most dramatically with C. parapsilosis, which is responsible for 10–25% of Candida infections of neonates and intensive care patients. C. parapsilosis infections are associated with the use of medical devices, especially central venous catheters, parenteral nutrition administration, and exposure to healthcare workers [21, 22].
In addition to traditional fungal culture methods to identify C. parapsilosis, molecular diagnosis based on nucleic acid detection is gaining importance, in particular, isothermal amplification technology and real-time qPCR technology. Isothermal amplification techniques can be divided into two categories based on the purpose of amplification: specific amplification and non-specific amplification. Besides RPA, LAMP, NASBA, SPIA, SDA, HDA, and RCA are also included. Among these amplification methods, only RPA is the only one that can automate isothermal amplification, while the others require DNA annealing and other necessary initial operations to achieve isothermal amplification, so RPA has a significant advantage in practical applications [14]. Real-time qPCR is specific and sensitive for large-scale detection of nucleic acids, especially for detection of novel coronaviruses, which are currently prevalent worldwide [23, 24]. RPA is tolerant of complex templates. Genomic DNA released by boiling method can be effectively detected. Release of DNA is more efficient with gram negative bacteria vs gram positive bacteria. Because the LOD of the RPA-LFS system was 5.85 × 103 copies/50 µl per reaction, the release of DNA of gram positive bacteria could not potentially impact the result of the assay. Due to the large-scale screening of nucleic acids conducted in various locations, the demand for laboratory testing by trained professionals continues to increase. In addition, most screening methods require expensive instruments for nucleic acid extraction and amplification. Moreover, aerosol contamination is a potential problem associated with testing of nucleic acids in a laboratory setting.
RPA is an effective complement to qPCR for testing of nucleic acids and can be used for home testing of patients or on-site testing in harsh conditions. Notably, this method allows for exponential amplification of nucleic acids at 37 °C without the need for expensive thermal sequencing instruments and professional laboratory personnel. Enzyme-linked immunosorbent assays are suitable for in vitro quantitative detection of proteins as markers of pathogens in cell cultures, serum, plasma, and other biological fluids. The double antibody sandwich method to detect antigenic proteins employs a microtiter plate with wells coated with a high-affinity biotinylated antibody for detection of the sample or standard. The presence of the target protein in the sample is detected by a colorimetric reaction with a substrate solution that is terminated by the addition of a termination solution [25]. However, this method does not allow effective detection at the early stage of disease, thus delaying prognosis and threatening the health of the patient. To compensate for this shortcoming, RPA is a highly sensitive, rapid, specific, and portable method for detection of C. parapsilosis.
Molecular detection techniques require the selection of diagnostic amplification targets to effectively detect specific species. Many studies have evaluated various methods for detection of C. parapsilosis. In the present study, a primer–probe set was designed to target FKS2 of C. parapsilosis. The results showed that there was no significant effect on the LOD for base mismatching and that the RPA-LFS assay accurately detected C. parapsilosis. The LOD of the RPA-LFS assay was 5.85 × 103 copies, which was more sensitive than qPCR at 102–103 copies per reaction. In addition, the RPA-LFS assay for detection of C. parapsilosis is simple and rapid, as the assay can be performed in 30 min or less. In addition, this method requires an isothermal temperature of only 37 °C, whereas PCR, qPCR, and loop-mediated isothermal amplification require temperature control equipment and relatively long reaction times. Evaluation of clinical samples showed that the RPA-LFS assay was specific. Testing of samples from different patients showed that the results of the RPA-LFS and qPCR assays were comparable, demonstrating that the RPA-LFS assay is a rapid and useful alternative. The detection system we have established for C. parapsilosis is consistent with many literature reports, featuring fast, portable, and instrument-independent characteristics. However, there is also a risk of aerosol contamination in the laboratory, which can be largely avoided by home testing.
Conclusions
The RPA-LFS assay developed in this study is a rapid highly specific and sensitive molecular technique for diagnosis of C. parapsilosis infection. Importantly, the test results are available within 40 min without the use of expensive instruments or trained laboratory personnel. Since testing can be conducted on-site, the proposed assay is useful for rapid detection of Candida spp. The established RPA-LFS assay is simple, rapid, and accurate, does not require laboratory facilities, and can be combined with a minimal and rapid DNA extraction method for home detection of C. parapsilosis infections for timely diagnosis to facilitate early treatment.
Data Availability
The data presented in this study are included in the article. Further inquiries should be directed to the corresponding authors.
References
Yacoub A, Moreland S, Jani D, Nanjappa S, Quilitz R, Carraway S, Sandin R, Greene J (2016) Candidemia in cancer patients: a retrospective analysis at a cancer center from 2001 to 2014. Infect Dis Clin Pract 24(5):273–277
Toth R, Toth A, Vagvolgyi C, Gacser A (2017) Candida parapsilosis secreted lipase as an important virulence factor. Curr Protein Pept Sci 18(10):1043–1049. https://doi.org/10.2174/1389203717666160813163054
Tóth R, Nosek J, Mora-Montes HM, Gabaldon T, Bliss JM, Nosanchuk JD, Turner SA, Butler G, Vágvölgyi C, Gácser A (2019) Candida parapsilosis: from genes to the bedside. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00111-18
Mesini A, Mikulska M, Giacobbe DR, Del Puente F, Gandolfo N, Codda G, Orsi A, Tassinari F, Beltramini S, Marchese A, Icardi G, Del Bono V, Viscoli C (2020) Changing epidemiology of candidaemia: increase in fluconazole-resistant Candida parapsilosis. Mycoses 63(4):361–368. https://doi.org/10.1111/myc.13050. (Epub 2020 Feb 20)
Guinea J, Mezquita S, Gómez A, Padilla B, Zamora E, Sánchez-Luna M, Sánchez-Carrillo C, Muñoz P, Escribano P (2021) Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med Mycol. https://doi.org/10.1093/mmy/myab068
Neji S, Trabelsi H, Hadrich I, Cheikhrouhou F, Sellami H, Makni F, Ayadi A (2017) Molecular study of the Candida parapsilosis complex in Sfax. Tunisia Med Mycol 55(2):137–144. https://doi.org/10.1093/mmy/myw063
Al-Tekreeti A, Al-Halbosiy M, Dheeb B, Hashim A, Al-Zuhairi A, Mohammad F (2018) Molecular identification of clinical Candida isolates by simple and randomly amplified polymorphic DNA-PCR. Arab J Sci Eng 43(1):163–170. https://doi.org/10.1007/s13369-017-2762-1
Souza A, Ferreira R, Gonçalves S, Quindós G, Eraso E, Bizerra F, Briones M, Colombo A (2012) Accurate identification of Candida parapsilosis (sensu lato) by use of mitochondrial DNA and real-time PCR. J Clin Microbiol 50(7):2310–2314. https://doi.org/10.1128/JCM.00303-12
Feng X, Wu Z, Ling B, Pan S, Liao W, Pan W, Yao Z (2014) Identification and differentiation of Candida parapsilosis complex species by use of exon-primed intron-crossing PCR. J Clin Microbiol 52(5):1758–1761. https://doi.org/10.1128/JCM.00105-14
Ferreira L, Sánchez-Juanes F, Porras-Guerra I, García-García M, García-Sánchez J, González-Buitrago J, Muñoz-Bellido J (2011) Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Microbiol Infect 17(4):546–551. https://doi.org/10.1111/j.1469-0691.2010.03257.x
Fallahi S, Babaei M, Rostami A, Mirahmadi H, Arab-Mazar Z, Sepahvand A (2020) Diagnosis of Candida albicans: conventional diagnostic methods compared to the loop-mediated isothermal amplification (LAMP) assay. Arch Microbiol 202(2):275–282. https://doi.org/10.1007/s00203-019-01736-7
Huang C, Huang P, Yao J, Li Z, Weng L, Guo X (2019) Pooled analysis of nuclear acid sequence-based amplification for rapid diagnosis of Mycoplasma pneumoniae infection. J Clin Lab Anal 33(5):e22879. https://doi.org/10.1002/jcla.22879
Carinelli S, Kühnemund M, Nilsson M, Pividori M (2017) Yoctomole electrochemical genosensing of Ebola virus cDNA by rolling circle and circle to circle amplification. Biosens Bioelectron 93:65–71. https://doi.org/10.1016/j.bios.2016.09.099
Yang H, Wang Y, Yang Q, Fan H, Wang L, Zhang T, Li Z, Liu G, Zhao P, Wu H (2021) A rapid and sensitive detection method for Pseudomonas aeruginosa using visualized recombinase polymerase amplification and lateral flow strip technology. Front Cell Infect Microbiol 11:698929. https://doi.org/10.3389/fcimb.2021.698929
Jeong Y, Park K, Kim D (2009) Isothermal DNA amplification in vitro: the helicase-dependent amplification system. Cellular Mol Life Sci: CMLS 66(20):3325–3336. https://doi.org/10.1007/s00018-009-0094-3
Zhang M, Li R, Ling L (2017) Homogenous assay for protein detection based on proximity DNA hybridization and isothermal circular strand displacement amplification reaction. Anal Bioanal Chem 409(16):4079–4085. https://doi.org/10.1007/s00216-017-0356-0
Piepenburg O, Williams C, Stemple D, Armes N (2006) DNA detection using recombination proteins. PLoS Biol. 4(7):e204. https://doi.org/10.1371/journal.pbio.0040204
Lee H, Yi S, Kwon J, Choi J, Lee D, Lee S, Shin Y (2021) Rapid and highly sensitive pathogen detection by real-time DNA monitoring using a nanogap impedimetric sensor with recombinase polymerase amplification. Biosens Bioelectron 179:113042. https://doi.org/10.1016/j.bios.2021.113042
Yang X, Xie J, Hu S, Zhan W, Duan L, Chen K, Zhang C, Yin A, Luo M (2020) Rapid and visual detection of enterovirus using recombinase polymerase amplification combined with lateral flow strips. Sens Actuators B: Chem. 311:127903. https://doi.org/10.1016/j.snb.2020.127903
Wang K, Huo L, Li Y, Zhu L, Wang Y, Wang L (2022) Establishment of a rapid diagnosis method for Candida glabrata based on the ITS2 gene using recombinase polymerase amplification combined with lateral flow strips. Front Cell Infect Microbiol. 12:953302. https://doi.org/10.3389/fcimb.2022.953302
Arikan K, Arıkan-Akdaglı S, Kara A (2019) 183. Candidaemia in children and importance of central venous catheter removal. Open Forum Infect Dis 6(Supplement 2):S112–S113. https://doi.org/10.1093/ofid/ofz360.258
Goemaere B, Becker P, Van Wijngaerden E, Maertens J, Spriet I, Hendrickx M, Lagrou K (2018) Increasing candidaemia incidence from 2004 to 2015 with a shift in epidemiology in patients preexposed to antifungals. Mycoses 61(2):127–133. https://doi.org/10.1111/myc.12714
Kim H, Schuck A, Lee S, Lee Y, Kang M, Kim Y (2021) Sensitive electrochemical biosensor combined with isothermal amplification for point-of-care COVID-19 tests. Biosens Bioelectron 182:113168. https://doi.org/10.1016/j.bios.2021.113168
Shelite T, Uscanga-Palomeque AC, Castellanos-Gonzalez A, Melby PC, Travi BL (2021) Isothermal recombinase polymerase amplification-lateral flow detection of SARS-CoV-2, the etiological agent of COVID-19. J Virological Methods. 296:114227. https://doi.org/10.1016/j.jviromet.2021.114227
Tsurusawa N, Chang J, Namba M, Makioka D, Yamura S, Iha K, Kyosei Y, Watabe S, Yoshimura T (2021) Modified ELISA for ultrasensitive diagnosis. J Clin Med 10(21):5197. https://doi.org/10.3390/jcm10215197
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding
This study was supported by grants from the Zhenjiang Key R&D Program (Social Development) (Grant No. SSH20220140238), the Lianyungang General Health Science and Technology Project in 2022 (Grant No. 202222).
Author information
Authors and Affiliations
Contributions
LZ, KW, and CC designed the experiments and wrote the manuscript. BZ and KW collected the clinical samples. BZ, LW, and YYL performed the main experiments. BZ analyzed the data. All authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare that they have no competing interests.
Ethics Approval
The study protocol was approved by the Medical Ethics Committee of the Second People’s Hospital of Lianyungang City (Lianyungang, Jiangsu, China) (permit number 2020013) and informed consent was obtained from patients prior to collection of clinical samples. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, B., Wang, L., Lu, Y. et al. Recombinase Polymerase Amplification Assay with Lateral Flow Strips for Rapid Detection of Candidiasis Due to Candida parapsilosis. Curr Microbiol 80, 217 (2023). https://doi.org/10.1007/s00284-023-03318-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03318-9