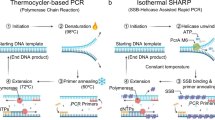
Abstract
Since the development of polymerase chain reaction, amplification of nucleic acids has emerged as an elemental tool for molecular biology, genomics, and biotechnology. Amplification methods often use temperature cycling to exponentially amplify nucleic acids; however, isothermal amplification methods have also been developed, which do not require heating the double-stranded nucleic acid to dissociate the synthesized products from templates. Among the several methods used for isothermal DNA amplification, the helicase-dependent amplification (HDA) is discussed in this review with an emphasis on the reconstituted DNA replication system. Since DNA helicase can unwind the double-stranded DNA without the need for heating, the HDA system provides a very useful tool to amplify DNA in vitro under isothermal conditions with a simplified reaction scheme. This review describes components and detailed aspects of current HDA systems using Escherichia coli UvrD helicase and T7 bacteriophage gp4 helicase with consideration of the processivity and efficiency of DNA amplification.





Similar content being viewed by others
References
Mullis KB, Faloona FA (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 155:335–350
Mullis KB (1990) The unusual origin of the polymerase chain reaction. Sci Am 262:56–61, 55–64
Mullis KB (1990) Target amplification for DNA analysis by the polymerase chain reaction. Ann Biol Clin (Paris) 48:579–582
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 24:476–480
Moore P (2005) PCR: replicating success. Nature 435:235–238
Brisson-Noel A, Gicquel B, Lecossier D, Levy-Frebault V, Nassif X, Hance AJ (1989) Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet 2:1069–1071
Lisby G, Dessau R (1994) Construction of a DNA amplification assay for detection of Legionella species in clinical samples. Eur J Clin Microbiol Infect Dis 13:225–231
Bernet C, Garret M, de Barbeyrac B, Bebear C, Bonnet J (1989) Detection of mycoplasma pneumoniae by using the polymerase chain reaction. J Clin Microbiol 27:2492–2496
Holland SM, Gaydos CA, Quinn TC (1990) Detection and differentiation of Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae by DNA amplification. J Infect Dis 162:984–987
Victor T, du Toit R, van Zyl J, Bester AJ, van Helden PD (1991) Improved method for the routine identification of toxigenic Escherichia coli by DNA amplification of a conserved region of the heat-labile toxin A subunit. J Clin Microbiol 29:158–161
Hoshina S, Kahn SM, Jiang W, Green PH, Neu HC, Chin N, Morotomi M, LoGerfo P, Weinstein IB (1990) Direct detection and amplification of Helicobacter pylori ribosomal 16S gene segments from gastric endoscopic biopsies. Diagn Microbiol Infect Dis 13:473–479
Duggan DB, Ehrlich GD, Davey FP, Kwok S, Sninsky J, Goldberg J, Baltrucki L, Poiesz BJ (1988) HTLV-I-induced lymphoma mimicking Hodgkin’s disease: diagnosis by polymerase chain reaction amplification of specific HTLV-I sequences in tumor DNA. Blood 71:1027–1032
Holodniy M, Katzenstein DA, Sengupta S, Wang AM, Casipit C, Schwartz DH, Konrad M, Groves E, Merigan TC (1991) Detection and quantification of human immunodeficiency virus RNA in patient serum by use of the polymerase chain reaction. J Infect Dis 163:862–866
Wang JY, Lee LN, Lai HC, Hsu HL, Jan IS, Yu CJ, Hsueh PR, Yang PC (2007) Performance assessment of the Capilia TB assay and the BD ProbeTec ET system for rapid culture confirmation of Mycobacterium tuberculosis. Diagn Microbiol Infect Dis 59:395–399
Piersimoni C, Scarparo C, Piccoli P, Rigon A, Ruggiero G, Nista D, Bornigia S (2002) Performance assessment of two commercial amplification assays for direct detection of Mycobacterium tuberculosis complex from respiratory and extrapulmonary specimens. J Clin Microbiol 40:4138–4142
Martro E, Garcia-Sierra N, Gonzalez V, Saludes V, Matas L, Ausina V (2009) Evaluation of an automated nucleic acid extractor for hepatitis C viral load quantification. J Clin Microbiol 47:811–813
Chan DJ, Ray JE, McNally L, Batterham M, Smith DE (2008) Correlation between HIV-1 RNA load in blood and seminal plasma depending on antiretroviral treatment status, regimen and penetration of semen by antiretroviral drugs. Curr HIV Res 6:477–484
Robuffo I, Fazii P, Rulli A, Di Nicola M, Toniato E, Di Rienzo M, Cosentino L, Gambi A, Castellani ML, Martinotti S (2008) Upgraded diagnostic value of Gen-Probe PACE 2 assay for detection of Chlamydia trachomatis infection. J Biol Regul Homeost Agents 22:253–261
Levett PN, Brandt K, Olenius K, Brown C, Montgomery K, Horsman GB (2008) Evaluation of three automated nucleic acid amplification systems for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in first-void urine specimens. J Clin Microbiol 46:2109–2111
Andras SC, Power JB, Cocking EC, Davey MR (2001) Strategies for signal amplification in nucleic acid detection. Mol Biotechnol 19:29–44
Vincent M, Xu Y, Kong H (2004) Helicase-dependent isothermal DNA amplification. EMBO Rep 5:795–800
Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP (1992) Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res 20:1691–1696
Fire A, Xu SQ (1995) Rolling replication of short DNA circles. Proc Natl Acad Sci USA 92:4641–4645
Demidov VV (2002) Rolling-circle amplification in DNA diagnostics: the power of simplicity. Expert Rev Mol Diagn 2:542–548
Runyon GT, Lohman TM (1989) Escherichia coli helicase II (uvrD) protein can completely unwind fully duplex linear and nicked circular DNA. J Biol Chem 264:17502–17512
Abdel-Monem M, Hoffmann-Berling H (1976) Enzymic unwinding of DNA. 1: purification and characterization of a DNA-dependent ATPase from Escherichia coli. Eur J Biochem 65:431–440
Lohman TM, Bjornson KP (1996) Mechanisms of helicase-catalyzed DNA unwinding [Review]. Annu Rev Biochem 65:169–214
Matson SW, Kaiser-Rogers KA (1990) DNA helicases. [Review] [306 refs]. Annu Rev Biochem 59:289–329
Patel SS, Picha KM (2000) Structure and function of hexameric helicases. Annu Rev Biochem 69:651–697
Patel SS, Donmez I (2006) Mechanisms of helicases. J Biol Chem 281:18265–18268
Kadare G, Haenni AL (1997) Virus-encoded RNA helicases. J Virol 71:2583–2590
Borowski P, Lang M, Niebuhr A, Haag A, Schmitz H, Schulze zur Wiesch J, Choe J, Siwecka MA, Kulikowski T (2001) Inhibition of the helicase activity of HCV NTPase/helicase by 1-beta-D-ribofuranosyl-1, 2, 4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP). Acta Biochim Pol 48:739–744
Borowski P, Lang M, Haag A, Schmitz H, Choe J, Chen HM, Hosmane RS (2002) Characterization of imidazo[4, 5-d]pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: evidence for activity on the level of substrate and/or enzyme. Antimicrob Agents Chemother 46:1231–1239
Borowski P, Lang M, Haag A, Baier A (2007) Tropolone and its derivatives as inhibitors of the helicase activity of hepatitis C virus nucleotide triphosphatase/helicase. Antivir Chem Chemother 18:103–109
Kwong AD, Rao BG, Jeang KT (2005) Viral and cellular RNA helicases as antiviral targets. Nat Rev Drug Discov 4:845–853
Hickman AB, Dyda F (2005) Binding and unwinding: SF3 viral helicases. Curr Opin Struct Biol 15:77–85
Gwack Y, Kim DW, Han JH, Choe J (1997) DNA helicase activity of the hepatitis C virus nonstructural protein 3. Eur J Biochem 250:47–54
Gorbalenya AE, Koonin EV (1993) Helicases: amino acidsequence comparisons and structure-function relationships. Curr Opin Struct Biol 3:419–429
Jennings TA, Mackintosh SG, Harrison MK, Sikora D, Sikora B, Tackett AJ, Cameron CE, Raney KD (2009) NS3 helicase from the hepatitis C Virus can function as a monomer or oligomer depending on enzyme and substrate concentrations. J Biol Chem 284:4806–4814
Xu HQ, Deprez E, Zhang AH, Tauc P, Ladjimi MM, Brochon JC, Auclair C, Xi XG (2003) The Escherichia coli RecQ helicase functions as a monomer. J Biol Chem 278:34925–34933
Nanduri B, Byrd AK, Eoff RL, Tackett AJ, Raney KD (2002) Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc Natl Acad Sci USA 99:14722–14727
Levin MK, Wang YH, Patel SS (2004) The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J Biol Chem 279:26005–26012
Byrd AK, Raney KD (2005) Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry 44:12990–12997
Tackett AJ, Chen Y, Cameron CE, Raney KD (2005) Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J Biol Chem 280:10797–10806
Donmez I, Patel SS (2006) Mechanisms of a ring shaped helicase. Nucleic Acids Res 34:4216–4224
Picha KM, Ahnert P, Patel SS (2000) DNA binding in the central channel of bacteriophage T7 helicase-primase is a multistep process; nucleotide hydrolysis is not required. Biochemistry 39:6401–6409
Picha KM, Patel SS (1998) Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+: the presence of dTTP is sufficient for hexamer formation and DNA binding. J Biol Chem 273:27315–27319
Kornberg A, Baker TA (1992) DNA replication. Freeman, New York
Kornberg A (1988) DNA replication. [Review] [14 refs]. Biochim Biophys Acta 951:235–239
Eckert KA, Kunkel TA (1990) High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res 18:3739–3744
Eckert KA, Kunkel TA (1991) DNA polymerase fidelity and the polymerase chain reaction. Genome Res 1:17–24
Andre P, Kim A, Khrapko K, Thilly WG (1997) Fidelity and mutational spectrum of Pfu DNA polymerase on a human mitochondrial DNA sequence. Genome Res 7:843–852
Cline J, Braman JC, Hogrefe HH (1996) PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res 24:3546–3551
Temin HM, Mizutani S (1970) RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211–1213
Baltimore D (1970) RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226:1209–1211
Joyce CM, Grindley ND (1983) Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc Natl Acad Sci USA 80:1830–1834
Jacobsen H, Klenow H, Overgaard-Hansen K (1974) The N-terminal amino-acid sequences of DNA polymerase I from Escherichia coli and of the large and the small fragments obtained by a limited proteolysis. Eur J Biochem 45:623–627
Krieg PA, Melton DA (1984) Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res 12:7057–7070
Schenborn ET, Mierendorf RC Jr (1985) A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res 13:6223–6236
Lohman TM, Bujalowski W, Overman LB (1988) E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends Biochem Sci 13:250–255
Lohman TM, Bujalowski W, Overman LB, Wei TF (1988) Interactions of the E. coli single strand binding (SSB) protein with ss nucleic acids: binding mode transitions and equilibrium binding studies. Biochem Pharmacol 37:1781–1782
Lohman TM, Overman LB, Datta S (1986) Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol 187:603–615
Weiner JH, Bertsch LL, Kornberg A (1975) The deoxyribonucleic acid unwinding protein of Escherichia coli: properties and functions in replication. J Biol Chem 250:1972–1980
Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34:4126–4137
Zou Y, Liu Y, Wu X, Shell SM (2006) Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol 208:267–273
Lohman TM, Ferrari ME (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and co-operativities. Annu Rev Biochem 63:527–570
Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43:289–318
Matson SW (1986) Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′–5′ direction. J Biol Chem 261:10169–10175
Lahue EE, Matson SW (1988) Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J Biol Chem 263:3208–3215
Maluf NK, Fischer CJ, Lohman TM (2003) A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol 325:913–935
An L, Tang W, Ranalli TA, Kim HJ, Wytiaz J, Kong H (2005) Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J Biol Chem 280:28952–28958
Hall MC, Jordan JR, Matson SW (1998) Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J 17:1535–1541
Mechanic LE, Frankel BA, Matson SW (2000) Escherichia coli MutL loads DNA helicase II onto DNA. J Biol Chem 275:38337–38346
Matson SW, Robertson AB (2006) The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res 34:4089–4097
Andresen D, von Nickisch-Rosenegk M, Bier FF (2009) Helicase dependent OnChip-amplification and its use in multiplex pathogen detection. Clin Chim Acta 403:244–248
Gill P, Alvandi AH, Abdul-Tehrani H, Sadeghizadeh M (2008) Colorimetric detection of Helicobacter pylori DNA using isothermal helicase-dependent amplification and gold nanoparticle probes. Diagn Microbiol Infect Dis 62:119–124
Gill P, Amini M, Ghaemi A, Shokouhizadeh L, Abdul-Tehrani H, Karami A, Gilak A (2007) Detection of Helicobacter pylori by enzyme-linked immunosorbent assay of thermophilic helicase-dependent isothermal DNA amplification. Diagn Microbiol Infect Dis 59:243–249
Goldmeyer J, Kong H, Tang W (2007) Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid RNA detection. J Mol Diagn 9:639–644
Motre A, Li Y, Kong H (2008) Enhancing helicase-dependent amplification by fusing the helicase with the DNA polymerase. Gene 420:17–22
Xu Y, Kim HJ, Kays A, Rice J, Kong H (2006) Simultaneous amplification and screening of whole plasmids using the T7 bacteriophage replisome. Nucleic Acids Res 34:e98
Dunn JJ, Studier FW (1983) Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol 166:477–535
Richardson CC (1983) Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule [Review]. Cell 33:315–317
Studier FW (1972) Bacteriophage T7 [Review]. Science 176:367–376
Bernstein JA, Richardson CC (1988) A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc Natl Acad Sci USA 85:396–400
Bernstein JA, Richardson CC (1989) Characterization of the helicase and primase activities of the 63-kDa component of the bacteriophage T7 gene 4 protein. J Biol Chem 264:13066–13073
Rosenberg AH, Patel SS, Johnson KA, Studier FW (1992) Cloning and expression of gene 4 of bacteriophage T7 and creation and analysis of T7 mutants lacking the 4A primase/helicase or the 4B helicase. J Biol Chem 267:15005–15012
Patel SS, Hingorani MM (1993) Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J Biol Chem 268:10668–10675
Hingorani MM, Patel SS (1996) Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry 35:2218–2228
Hingorani MM, Patel SS (1993) Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry 32:12478–12487
Ahnert P, Patel SS (1997) Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J Biol Chem 272:32267–32273
Jeong YJ, Levin MK, Patel SS (2004) The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci USA 101:7264–7269
Matson SW, Richardson CC (1983) DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem 258:14009–14016
Ahnert P, Picha KM, Patel SS (2000) A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J 19:3418–3427
Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS (1995) Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci USA 92:3869–3873
Tabor S, Huber HE, Richardson CC (1987) Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem 262:16212–16223
Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC (2007) Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell 27:539–549
Kim YT, Tabor S, Bortner C, Griffith JD, Richardson CC (1992) Purification and characterization of the bacteriophage T7 gene 2.5 protein: a single-stranded DNA-binding protein. J Biol Chem 267:15022–15031
Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T (2001) Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc Natl Acad Sci USA 98:9557–9562
Nakai H, Richardson CC (1988) Leading and lagging strand synthesis at the replication fork of bacteriophage T7: distinct properties of T7 gene 4 protein as a helicase and primase. J Biol Chem 263:9818–9830
Kim YT, Tabor S, Churchich JE, Richardson CC (1992) Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J Biol Chem 267:15032–15040
Kim DE, Patel SS (2002) T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J Mol Biol 321:807–819
Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS (2005) DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature 435:370–373
Notarnicola SM, Mulcahy HL, Lee J, Richardson CC (1997) The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J Biol Chem 272:18425–18433
Li Y, Kim HJ, Zheng C, Chow WH, Lim J, Keenan B, Pan X, Lemieux B, Kong H (2008) Primase-based whole genome amplification. Nucleic Acids Res 36:e79
Kusakabe T, Richardson CC (1997) Gene 4 DNA primase of bacteriophage T7 mediates the annealing and extension of ribo-oligonucleotides at primase recognition sites. J Biol Chem 272:12446–12453
Tabor S, Richardson CC (1981) Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci USA 78:205–209
Benkovic SJ, Valentine AM, Salinas F (2001) Replisome-mediated DNA replication. Annu Rev Biochem 70:181–208
Nossal NG (1992) Protein–protein interactions at a DNA replication fork: bacteriophage T4 as a model [Review]. FASEB J 6:871–878
Young MC, Reddy MK, von Hippel PH (1992) Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry 31:8675–8690
Delagoutte E, von Hippel PH (2001) Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry 40:4459–4477
Kim S, Dallmann HG, McHenry CS, Marians KJ (1996) Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell 84:643–650
McHenry CS (2003) Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol Microbiol 49:1157–1165
Kim S, Dallmann HG, McHenry CS, Marians KJ (1996) Tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J Biol Chem 271:21406–21412
Galletto R, Jezewska MJ, Bujalowski W (2004) Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol 343:83–99
Bujalowski W, Jezewska MJ (1995) Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA: the nucleic acid does not wrap around the protein hexamer. Biochemistry 34:8513–8519
Barcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher M, Carazo JM (2001) The DnaB.DnaC complex: a structure based on dimers assembled around an occluded channel. EMBO J 20:1462–1468
Xi J, Zhang Z, Zhuang Z, Yang J, Spiering MM, Hammes GG, Benkovic SJ (2005) Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome. Biochemistry 44:7747–7756
Lohman TM, Tomko EJ, Wu CG (2008) Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9:391–401
Zhang W, Dillingham MS, Thomas CD, Allen S, Roberts CJ, Soultanas P (2007) Directional loading and stimulation of PcrA helicase by the replication initiator protein RepD. J Mol Biol 371:336–348
Pang PS, Jankowsky E, Planet PJ, Pyle AM (2002) The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J 21:1168–1176
Acknowledgments
This work was supported by a grant (20080401034026) from the BioGreen 21 Program, Rural Development Administration, Republic of Korea, a grant from Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (A080123), a grant (10032113) from Industrial Technology Development, Ministry of Knowledge Economy, and a grant from the Korea Science & Engineering Foundation (KOSEF) through a general research grant (R01-2008-000-20301-0). K. Park is supported by the second stage of Brain Korea 21. Y.-J. Jeong was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2008-313-C00531).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jeong, YJ., Park, K. & Kim, DE. Isothermal DNA amplification in vitro: the helicase-dependent amplification system. Cell. Mol. Life Sci. 66, 3325–3336 (2009). https://doi.org/10.1007/s00018-009-0094-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0094-3