Abstract
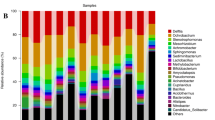
Chlorpyrifos (CPF), a common organophosphorus pesticide, is extensively used in agricultural practices. However, we lack sound evidence for the linkage between soil microbial diversity, soil function, and plant biomass under the influence of CPF, which prevents us from assessing the actual impact of CPF on agricultural production. In this study, we used high-throughput sequencing to test the effects of CPF on soil microbial diversity, soil function, and cotton biomass in indoor pot experiments. The use of CPF leads to a significant reduction in cotton biomass until the concentration of CPF used reaches 15 mg kg−1, and the cotton biomass is no longer significantly reduced. Compared with the original soil, the alpha-diversity of bacteria, which was significantly linearly related to cotton biomass, was significantly decreased when the soil was treated with 15 mg kg−1 CPF. Affected by CPF, the overall soil microbial composition has changed significantly. Acidobacteria, Nitrospirae, Planctomycetes, and Actinobacteria were significantly regulated after CPF treatment. Correspondingly, key soil functions, including nitrogen metabolism and iron (III) transporter, have been significantly down-regulated. The reduction of nitrogen and Fe3+ should deprive the cotton of essential nutrients during the short crop cycle and thus affect cotton biomass. Our study provides experimental evidence that CPF affects cotton rhizosphere soil microbial diversity, the relative content of key bacterial genera, and soil function, which shows that it has an important impact on plant biomass, and provides a reference for studying the actual impact of CPF on the environment and agricultural production.





Similar content being viewed by others
Data Availability
The data presented in this study are openly available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession number PRJNA662843 and Supplementary Material of this article.
References
Babina K, Dollard M, Pilotto L, Edwards JW (2012) Environmental exposure to organophosphorus and pyrethroid pesticides in South Australian preschool children: a cross sectional study. Environ Int 48:109–120. https://doi.org/10.1016/j.envint.2012.07.007
Huang X, Cui H, Duan W (2020) Ecotoxicity of chlorpyrifos to aquatic organisms: a review. Ecotoxicol Environ Saf. https://doi.org/10.1016/j.ecoenv.2020.110731
Bishnu A, Chakrabarti K, Chakraborty A, Saha T (2009) Pesticide residue level in tea ecosystems of Hill and Dooars regions of West Bengal, India. Environ Monit Assess 149(1–4):457–464. https://doi.org/10.1007/s10661-008-0222-9
Yuan Y, Chen C, Zheng C, Wang X, Yang G, Wang Q, Zhang Z (2014) Residue of chlorpyrifos and cypermethrin in vegetables and probabilistic exposure assessment for consumers in Zhejiang Province, China. Food Control 36(1):63–68. https://doi.org/10.1016/j.foodcont.2013.08.008
Parveen Z, Khuhro MI, Rafiq N, Kausar N (2004) Evaluation of multiple pesticide residues in apple and citrus fruits, 1999–2001. Bull Environ Contam Toxicol 73(2):312–318. https://doi.org/10.1007/s00128-004-0429-6
John EM, Shaike JM (2015) Chlorpyrifos: pollution and remediation. Environ Chem Lett 13(3):269–291. https://doi.org/10.1007/s10311-015-0513-7
Fang H, Yu Y, Chu X, Wang X, Yang X, Yu J (2009) Degradation of chlorpyrifos in laboratory soil and its impact on soil microbial functional diversity. J Environ Sci 21(3):380–386. https://doi.org/10.1016/s1001-0742(08)62280-9
Tipayno S, Kim C-G, Sa T (2012) T-RFLP analysis of structural changes in soil bacterial communities in response to metal and metalloid contamination and initial phytoremediation. Appl Soil Ecol 61:137–146. https://doi.org/10.1016/j.apsoil.2012.06.001
Gilani RA, Rafique M, Rehman A, Munis MFH, Rehman SU, Chaudhary HJ (2016) Biodegradation of chlorpyrifos by bacterial genus Pseudomonas. J Basic Microbiol 56(2):105–119. https://doi.org/10.1002/jobm.201500336
Baskaran S, Kookana RS, Naidu R (2003) Contrasting behaviour of chlorpyrifos and its primary metabolite, TCP (3,5,6-trichloro-2-pyridinol), with depth in soil profiles. Aust J Soil Res 41(4):749–760. https://doi.org/10.1071/sr02062
Menon P, Gopal M, Parsad R (2005) Effects of chlorpyrifos and quinalphos on dehydrogenase activities and reduction of Fe3+ in the soils of two semi-arid fields of tropical India. Agric Ecosyst Environ 108(1):73–83. https://doi.org/10.1016/j.agee.2004.12.008
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17(8):478–486. https://doi.org/10.1016/j.tplants.2012.04.001
Bender SF, Wagg C, van der Heijden MGA (2016) An Underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31(6):440–452. https://doi.org/10.1016/j.tree.2016.02.016
Singh BK, Quince C, Macdonald CA, Khachane A, Thomas N, Abu Al-Soud W, Sorensen SJ, He Z, White D, Sinclair A, Crooks B, Zhou J, Campbell CD (2014) Loss of microbial diversity in soils is coincident with reductions in some specialized functions. Environ Microbiol 16(8):2408–2420. https://doi.org/10.1111/1462-2920.12353
Pandey S, Singh DK (2004) Total bacterial and fungal population after chlorpyrifos and quinalphos treatments in groundnut (Arachis hypogaea L.) soil. Chemosphere 55(2):197–205. https://doi.org/10.1016/j.chemosphere.2003.10.014
Adak T, Munda S, Kumar U, Berliner J, Pokhare SS, Jambhulkar NN, Jena M (2016) Effect of elevated CO2 on chlorpyriphos degradation and soil microbial activities in tropical rice soil. Environ Monit Assess. https://doi.org/10.1007/s10661-016-5119-4
Sarnaik SS, Kanekar PP, Raut VM, Taware SP, Chavan KS, Bhadbhade BJ (2006) Effect of application of different pesticides to soybean on the soil microflora. J Environ Biol 27(2):423–426
Pozo C, Rodelas B, Gonzalez-Lopez J, Salmeron V, Martinez-Toledo MV (1995) Effect of chlorpyrifos on soil microbial activity. Environ Toxicol Chem 14(2):187–192. https://doi.org/10.1002/etc.5620140201
Shan M, Fang H, Wang X, Feng B, Chu XQ, Yu YL (2006) Effect of chlorpyrifos on soil microbial populations and enzyme activities. J Environ Sci 18(1):4–5
Sardar D, Kole RK (2005) Metabolism of chlorpyrifos in relation to its effect on the availability of some plant nutrients in soil. Chemosphere 61(9):1273–1280. https://doi.org/10.1016/j.chemosphere.2005.03.078
Chen QL, Ding J, Zhu YG, He JZ, Hu HW (2020) Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environ Int 140:7. https://doi.org/10.1016/j.envint.2020.105766
Guo A, Pan C, Ma J, Bao Y (2020) Linkage of antibiotic resistance genes, associated bacteria communities and metabolites in the wheat rhizosphere from chlorpyrifos-contaminated soil. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2020.140457
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488(7409):91–95. https://doi.org/10.1038/nature11336
Ruiz JM, Castilla N, Romero L (2000) Nitrogen metabolism in pepper plants applied with different bioregulators. J Agric Food Chem 48(7):2925–2929. https://doi.org/10.1021/jf990394h
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072. https://doi.org/10.1128/aem.03006-05
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu S-M, Peng S, Zhu X, Liu G, Liao X, Li Y, Yang H, Wang J, Lam T-W, Wang J (2015) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler (vol 1, 18, 2012). Gigascience. https://doi.org/10.1186/s13742-015-0069-2
Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL (2018) The soil-borne legacy. Cell 172(6):1178–1180. https://doi.org/10.1016/j.cell.2018.02.024
Parween T, Jan S, Mahmooduzzafar FT (2012) Evaluation of oxidative stress in Vigna radiata L. in response to chlorpyrifos. Int J Environ Sci Technol 9(4):605–612. https://doi.org/10.1007/s13762-012-0095-x
Bengtsson J (1998) Which species? What kind of diversity? Which ecosystem function? Some problems in studies of relations between biodiversity and ecosystem function. Appl Soil Ecol 10(3):191–199
Huang X, Cui H, Yang L, Lan T, Zhang J, Cai Z (2017) The microbial changes during the biological control of cucumber damping-off disease using biocontrol agents and reductive soil disinfestation. Biocontrol 62(1):97–109. https://doi.org/10.1007/s10526-016-9768-6
Tebo BM, Davis RE, Anitori RP, Conne LB, Schiffman P, Staudigel H (2015) Microbial communities in dark oligotrophic volcanic ice cave ecosystems of Mt. Erebus, Antarctica. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00179
Huang X-F, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM (2014) Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany. https://doi.org/10.1139/cjb-2013-0225
Lu L, Xing D, Ren ZJ (2015) Microbial community structure accompanied with electricity production in a constructed wetland plant microbial fuel cell. Bioresour Technol 195:115–121. https://doi.org/10.1016/j.biortech.2015.05.098
Gaonkar O, Nambi IM, Suresh Kumar G (2019) Biodegradation kinetics of dichlorvos and chlorpyrifos by enriched bacterial cultures from an agricultural soil. Biorem J 23(4):259–276. https://doi.org/10.1080/10889868.2019.1671791
Sasikala C, Jiwal S, Rout P, Ramya M (2012) Biodegradation of chlorpyrifos by bacterial consortium isolated from agriculture soil. World J Microbiol Biotechnol 28(3):1301–1308. https://doi.org/10.1007/s11274-011-0879-z
Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M (2015) Complete nitrification by Nitrospira bacteria. Nature 528(7583):504–509. https://doi.org/10.1038/nature16461
Kumar U, Berliner J, Adak T, Rath PC, Dey A, Pokhare SS, Jambhulkar NN, Panneerselvam P, Kumar A, Mohapatra SD (2017) Non-target effect of continuous application of chlorpyrifos on soil microbes, nematodes and its persistence under sub-humid tropical rice-rice cropping system. Ecotoxicol Environ Saf 135:225–235. https://doi.org/10.1016/j.ecoenv.2016.10.003
Racke KD, Laskowski DA, Schultz MR (1990) Resistance of chlorpyrifos to enhanced biodegradation in soil. J Agric Food Chem 38(6):1430
Xun WB, Li W, Xiong W, Ren Y, Liu YP, Miao YZ, Xu ZH, Zhang N, Shen QR, Zhang RF (2019) Diversity-triggered deterministic bacterial assembly constrains community functions. Nat Commun 10:10. https://doi.org/10.1038/s41467-019-11787-5
Lu C, Yang Z, Liu J, Liao Q, Ling W, Waigi MG, Odinga ES (2020) Chlorpyrifos inhibits nitrogen fixation in rice-vegetated soil containing Pseudomonas stutzeri A1501. Chemosphere. https://doi.org/10.1016/j.chemosphere.2020.127098
Bassey IY (2015) Germination and root nodule formation of soybean (Glycine max (L.) Merr.) in Ridomil and chlorpyriphos treated soil. Am J Environ Prot 4(1):17–22. https://doi.org/10.1648/j.ajep.20150401.12
Singh S, Gupta R, Sharma S (2015) Effects of chemical and biological pesticides on plant growth parameters and rhizospheric bacterial community structure in Vigna radiata. J Hazard Mater 291:102–110. https://doi.org/10.1016/j.jhazmat.2015.02.053
Singh S, Gupta R, Kumari M, Sharma S (2015) Nontarget effects of chemical pesticides and biological pesticide on rhizospheric microbial community structure and function in Vigna radiata. Environ Sci Pollut Res 22(15):11290–11300. https://doi.org/10.1007/s11356-015-4341-x
Walvekar VA, Bajaj S, Singh DK, Sharma S (2017) Ecotoxicological assessment of pesticides and their combination on rhizospheric microbial community structure and function of Vigna radiata. Environ Sci Pollut Res 24(20):17175–17186. https://doi.org/10.1007/s11356-017-9284-y
Prasertsup P, Ariyakanon N (2011) Removal of Chlorpyrifos by Water Lettuce (Pistia stratiotes L.) and Duckweed (Lemna minor L. ). Int J Phytorem 13(4):383–395. https://doi.org/10.1080/15226514.2010.495145
Ye ZH, Cheung KC, Wong MH (2003) Cadmium and nickel adsorption and uptake in cattail as affected by iron and manganese plaque on the root surface. Commun Soil Sci Plant Anal 34(19–20):2763–2778. https://doi.org/10.1081/css-120025202
Schatz A, Bugie E, Waksman SA (2005) Streptomycin, a substance exhibiting antibiotic activity against gram-positive and Gram-negative bacteria (vol 437, pg 3, 2005). Clin Orthop Rel Res 441:379–379
Acknowledgements
The authors would like to thank all the students who participated in this project.
Funding
This research was supported by the Shanghai Sailing Program (21YF1410100), and the National High-tech R&D Program of China (863 Program) (No. 2012AA101401).
Author information
Authors and Affiliations
Contributions
Conceptualization: XW, JW, and YW designed the experiments. XW, JC, and XZ performed the experiments. XW and WW analyzed the results and wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors hereby declare that they have no conflict of interest.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Wang, J., Wang, Y. et al. Changes in Microbial Diversity, Soil Function, and Plant Biomass of Cotton Rhizosphere Soil Under the Influence of Chlorpyrifos. Curr Microbiol 79, 323 (2022). https://doi.org/10.1007/s00284-022-03015-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03015-z