Abstract
According to the latest Newcastle disease virus (NDV) classification system, Iranian PPMV-1 isolates were classified as either XXI.1.1 or XXI.2 subgenotypes only. However, a few recent studies have suggested the possible existence of other Iranian PPMV-1 genotypes/subgenotypes. Recently, we isolated a PPMV-1 closely related to the African origin subgenotype VI.2.1.2 from an ill captive pigeon in a park aviary in central Tehran (Pg/IR/AMMM160/2019). This subgenotype had never been reported from Iran or neighboring countries. We also isolated a subgenotype VII.1.1 NDV (Pg/IR/AMMM117/2018), usually reported from non-pigeon birds in Iran. The nucleotide distance of AMMM117 was 1.0–2.5% compared to other Iranian subgenotypes VII.1.1 isolates. However, usually the same year VII.1.1 viruses that we isolate from Iranian poultry farms show negligible distances (0.0–0.5%). More isolates are required to study if this difference is due to subgenotype VII.1.1 being circulated and mutated in pigeons. Here, we also characterized two other isolates, namely Pg/IR/AMMM168/2019 and Pg/IR/MAM39/2017. The latter is the first Iranian subgenotype XXI.1.1 to be featured in the NDV datasets of the international NDV consortium. We also investigated the phylogenetic relation of all the published Iranian pigeon-derived NDV to date and updated the grouping according to the latest classification system. We have concluded that at least six different groups of pigeon-derived NDV have been circulating in Iran since 1996, four of which have been reported from just one city over the last seven years. This study suggests that the Iranian pigeon-origin NDV have been more diverse than the Iranian poultry-derived NDV in recent years.



Similar content being viewed by others
Abbreviations
- Pigeon paramyxovirus serotype-1:
-
PPMV-1
- Newcastle disease virus:
-
NDV
- specific pathogen-free:
-
SPF
- hemagglutination assay:
-
HA
- hemagglutination inhibition assay:
-
HI
- Fusion:
-
F
- Basic Local Alignment Search Tool:
-
BLAST
- mean death time:
-
MDT
- intracerebral pathogenicity index:
-
ICBI
- coding sequence:
-
CDS
References
Dimitrov KM, Afonso CL, Yu Q, Miller PJ (2016) Newcastle disease vaccines-A solved problem or a continuous challenge? Vet Microbiol 206:126–136
Dimitrov KM, Abolnik C, Afonso CL, Albina E, Bahl J, Berg M, Briand FX, Brown IH, Choi KS, Chvala I, Diel DG, Durr PA, Ferreira HL, Fusaro A, Gil P, Goujgoulova GV, Grund C, Hicks JT, Joannis TM, Torchetti MK, Kolosov S, Lambrecht B, Lewis NS, Liu H, Liu H, McCullough S, Miller PJ, Monne I, Muller CP, Munir M, Reischak D, Sabra M, Samal SK, Servan de Almeida R, Shittu I, Snoeck CJ, Suarez DL, Van Borm S, Wang Z, Wong FYK (2019) Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol 74:103917. https://doi.org/10.1016/j.meegid.2019.103917
Dortmans JC, Koch G, Rottier PJ, Peeters BP (2011) A comparative infection study of pigeon and avian paramyxovirus type 1 viruses in pigeons: evaluation of clinical signs, virus shedding and seroconversion. Avian Pathol 40(2):125–130. https://doi.org/10.1080/03079457.2010.542131
Kommers GD, King DJ, Seal BS, Brown CC (2001) Virulence of pigeon-origin Newcastle disease virus isolates for domestic chickens. Avian Dis 45(4):906–921. https://doi.org/10.2307/1592870
Ferreira HL, Taylor TL, Dimitrov KM, Sabra M, Afonso CL, Suarez DL (2019) Virulent Newcastle disease viruses from chicken origin are more pathogenic and transmissible to chickens than viruses normally maintained in wild birds. Vet Microbiol 235:25–34. https://doi.org/10.1016/j.vetmic.2019.06.004
Czeglédi A, Ujvári D, Somogyi E, Wehmann E, Werner O, Lomniczi B (2006) Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res 120(1–2):36–48. https://doi.org/10.1016/j.virusres.2005.11.009
Karsunke J, Heiden S, Murr M, Karger A, Franzke K, Mettenleiter TC, Romer-Oberdorfer A (2019) W protein expression by Newcastle disease virus. Virus Res 263:207–216. https://doi.org/10.1016/j.virusres.2019.02.003
OIE (2012) Newcastle disease. In: Manual of diagnostic tests and vaccines for terrestrial animals, vol 1. 7th ed. edn. World Organisation for Animal Health, Paris, France, pp 555–573
Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL (2012) Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol 12(8):1770–1779. https://doi.org/10.1016/j.meegid.2012.07.012
Rezaei Far A, Peighambari SM, Pourbakhsh SA, Ashtari A, Soltani M (2017) Co-circulation of genetically distinct groups of avian paramyxovirus type 1 in pigeon Newcastle disease in Iran. Avian Pathol 46(1):36–43. https://doi.org/10.1080/03079457.2016.1203068
Soltani M, Peighambari SM, Pourbakhsh SA, Ashtari A, Rezaei Far A, Abdoshah M (2019) Molecular characterization of haemagglutinin-neuraminidase gene among virulent Newcastle disease viruses isolated in Iran. Iran J Vet Res 20(1):1–8
Mayahi M, Seyfi Abad Shapouri MR, Jafari RA, KhosraviFarsani M (2017) Characterization of isolated pigeon paramyxovirus-1 (PMV-1) and its pathogenicity in broiler chickens. Vet Res Forum 8(1):15–21
Mayahi M, Sefi Abad Shapouri MR, Jafary RA, KhosraviFarsani M (2015) Isolation and molecular diagnostic of pigeon paramyxovirus-1 from suspected pigeons to Newcastle disease in Ahvaz. Iran Iranian Vet J 11(2):102–112
Mayahi V, Esmaelizad M (2017) Molecular evolution and epidemiological links study of Newcastle disease virus isolates from 1995 to 2016 in Iran. Arch Virol 162(12):3727–3743. https://doi.org/10.1007/s00705-017-3536-5
Langroudi G, Hosseini H, Karimi V, Hashemzadeh M, Ghafouri A, Ghadikolaei B, VahedisM, (2014) Phylogenetic study based on the phosphoprotein gene of Iranian Newcastle disease viruses (NDV) isolates, 2010–2012. Iran J Vet Med 8(2):73–77
Ujvari D, Wehmann E, Kaleta E, Werner O, Savić V, Nagy E, Czifra G, Lomniczi B (2003) Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res 96:63–73. https://doi.org/10.1016/S0168-1702(03)00173-4
Molouki A, Mehrabadi MHF, Bashashati M, Akhijahani MM, Lim SHE, Hajloo SA (2019) NDV subgenotype VII(L) is currently circulating in commercial broiler farms of Iran, 2017–2018. Trop Anim Health Prod 51(5):1247–1252. https://doi.org/10.1007/s11250-019-01817-1
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Allahyari E, Allymehr M, Molouki A, FallahMehrabadi MH, Talebi A (2020) Molecular characterization and phylogenetic study of the fusion gene of Newcastle disease viruses isolated from broiler farms of Iran in 2018–2019. Bulg J Vet Med. https://doi.org/10.15547/bjvm.2020-0041
Aldous EW, Mynn JK, Banks J, Alexander DJ (2003) A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol 32(3):239–256. https://doi.org/10.1080/030794503100009783
Ellakany HF, Elbestawy AR, Abd El-Hamid HS, Zedan RE, Gado AR, Taha AE, Soliman MA, Abd El-Hack ME, Swelum AA, Saadeldin IM, Ba-Awadh H, Hussein EOS (2019) Role of pigeons in the transmission of avian avulavirus (Newcastle Disease-Genotype VIId) to chickens. Animals (Basel) 9(6):338. https://doi.org/10.3390/ani9060338
Dimitrov K, Bolotin V, Muzyka D, Goraichuk I, Solodiankin O, Gerilovych A, Stegniy B, Goujgoulova G, Silko N, Pantin-Jackwood M, Miller P, Afonso C (2016) Repeated isolation of virulent Newcastle disease viruses of sub-genotype VIId from backyard chickens in Bulgaria and Ukraine between 2002 and 2013. Arch Virol 161(12):3345–3353. https://doi.org/10.1007/s00705-016-3033-2
Ghalyanchilangeroudi A, Hosseini H, Jabbarifakhr M, FallahMehrabadi MH, Najafi H, Ghafouri SA, Mousavi FS, Ziafati Z, Modiri A (2018) Emergence of a virulent genotype VIIi of newcastle disease virus in Iran. Avian Pathol 47(5):509–519. https://doi.org/10.1080/03079457.2018.1495313
Sohrab V (1973) Newcastle disease in Iran. Bull Off Int Epiz 79(5–6):565–569
Kianizadeh M, Aini I, Omar AR, Yusoff K, Sahrabadi M, Kargar R (2002) Sequence and phylogenetic analysis of the fusion protein cleavage site of Newcastle disease virus field isolates from Iran. Acta Virol 46(4):247–251
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101(30):11030–11035. https://doi.org/10.1073/pnas.0404206101
Acknowledgements
This work was supported by Razi Institute under the grant number 1-18-18-062-97020.
Author information
Authors and Affiliations
Contributions
AM and MS designed and performed the experiments. MS and MHFM provided samples. EA and AM performed computer analysis. MMA and AA performed inoculation, HA and HI. MA performed MDT and ICPI. AM wrote the first draft of the manuscript. MS, SHEL, AG edited the manuscript. SAP and AS were involved in executing the study and providing laboratory space.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Research involving human participants and/or animals
The present study did not involve any human subject. Animal handling procedures were performed in line with the national animal welfare regulations. The Institutional Animal Care and Use Committee (IACUC) of Razi Vaccine and Serum Research Institute approved all animal experiments.
Informed consent
The present study did not involve any human subject.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary File 1
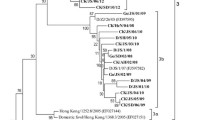
. The phylogenetic tree for all the Iranian pigeon-derived NDV made using the large dataset of the international NDV consortium. Six distinct groups were observed based on the sequence of the F gene. The evolutionary history was inferred by using the Maximum Likelihood method (bootstrap 1000) and General Time Reversible model [26]. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.1762)). This analysis involved 1789 nucleotide sequences. Evolutionary analyses were conducted in MEGA X [18]. All the isolates of this study are marked with ●. The file must be opened in MEGA X. Supplementary file 1 (MTSX 1395 KB)
Below is the link to the electronic supplementary material.
Supplementary File 2
. Estimates of evolutionary divergence between the F gene sequences of Iranian pigeon-derived isolates and all the sequences available in the large dataset of the NDV consortium. Analyses were conducted using the Maximum Composite Likelihood model [26] and a bootstrap 1000. The rate variation among sites was modeled with a gamma distribution (shape parameter=1). The analysis involved 1789 nucleotide sequences. The file must be viewed with text editor in MEGA X. Supplementary file 2 (ZIP 5830 KB)
Below is the link to the electronic supplementary material.
Supplementary File 3
. A phylogenetic tree for the seven complete F gene CDS of the Iranian pigeon-derived NDV. The tree was constructed using the pilot dataset of the NDV consortium. The evolutionary history was inferred by using the Maximum Likelihood method (bootstrap 500) and General Time Reversible model [26]. All the isolates of this study are marked with ●. Evolutionary analyses were conducted in MEGA X [18]. Supplementary file 3 (TIF 12889 KB)
Below is the link to the electronic supplementary material.
Supplementary File 4
. Estimates of evolutionary divergence between the seven Iranian pigeon-derived NDV and the isolates in the large dataset of the NDV consortium based on the complete F gene CDS. Analyses were conducted using the Maximum Composite Likelihood model [26] and a bootstrap 500. The file must be viewed using the text editor in MEGA X. Supplementary file 4 (ZIP 7844 KB)
Below is the link to the electronic supplementary material.
Supplementary File 5
. A FASTA file prepared using all the partial/complete F gene sequences of Iranian PPMV-1 and the pilot file of the NDV consortium. The genotype/subgenotype of all the isolates is shown in their title. An updated version of the file will be available in our Google Drive (see text for URL link). Supplementary file 5 (FAS 280 KB)
Rights and permissions
About this article
Cite this article
Molouki, A., Soltani, M., Akhijahani, M.M. et al. Circulation of at Least Six Distinct Groups of Pigeon-Derived Newcastle Disease Virus in Iran Between 1996 and 2019. Curr Microbiol 78, 2672–2681 (2021). https://doi.org/10.1007/s00284-021-02505-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02505-w