Abstract
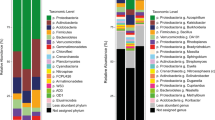
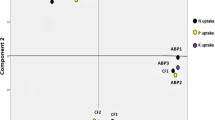
Plant growth reduction caused by osmotic stress, pathogens, and nutrient scarcity can be overcome by inoculation with plant growth-promoting rhizobacteria (PGPR). Knowing the effects of PGPR on the microbial community beyond those on plant growth can bring new options of soil microbiota management. The present study aimed to investigate the effect of inoculation with the newly described Pseudomonas aestus CMAA 1215T [a 1-aminocyclopropane-1-carboxylate (ACC) deaminase and glycine-betaine producer] on the rhizosphere bacterial community of Zea mays in natural (non-salinized) and saline soil. The bacterial community structure was assessed by sequencing the V6–V7 16S ribosomal RNA using the Ion Personal Genome Machine™. The non-metric multidimensional scaling (NMDS) of the OTU profile (ANOSIM P < 0.01) distinguishes all the treatments (with and without inoculation under saline and natural soils). Inoculated samples shared 1234 OTUs with non-inoculated soil. The most abundant classes in all samples were Alphaproteobacteria, Gammaproteobacteria, Actinobacteria, Acidobacteriia, Bacteroidia, Thermoleophilia, Verrucomicrobiae, Ktenodobacteria, and Bacilli. The inoculation, on the other hand, caused an increase in the abundance of the genera Bacillus, Bryobacter, Bradyrhizobium, “Candidatus Xiphinematobacter”, and “Candidatus Udaeobacter” independent of soil salinization. “Candidatus Udaeobacter” has the largest Mean Decrease in Gini Values with higher abundance on inoculated salted soil. In addition, Pseudomonas inoculation reduced the abundance of Gammaproteobacteria and Phycisphaerae. Understanding how inoculation modifies the bacterial community is essential to manage the rhizospheric microbiome to create a multi-inoculant approach and to understand its effects on ecological function.





Similar content being viewed by others
References
Da Rocha UN, Plugge CM, George I et al (2013) The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PLoS ONE 8:16–20. https://doi.org/10.1371/journal.pone.0082443
Shi S, Nuccio E, Herman DJ et al (2015) Successional trajectories of rhizosphere bacterial communities over consecutive seasons. MBio 6:e00746-e815. https://doi.org/10.1128/mBio.00746-15
Buée M, De BW, Martin F et al (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212. https://doi.org/10.1007/s11104-009-9991-3
Xu X, Passey T, Wei F et al (2015) Amplicon-based metagenomics identified candidate organisms in soils that caused yield decline in strawberry. Hortic Res 2:15022. https://doi.org/10.1038/hortres.2015.22
Jha CK, Saraf M (2015) Plant growth promoting Rhizobacteria (PGPR): a review. J Agric Res Dev 5:108–119. https://doi.org/10.13140/RG.2.1.5171.2164
Glick BR, Penrose DM, Li J (1998) A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J Theor Biol i 190:63–68
Chihaoui S-A, Trabelsi D, Jdey A et al (2015) Inoculation of Phaseolus vulgaris with the nodule-endophyte Agrobacterium sp. 10C2 affects richness and structure of rhizosphere bacterial communities and enhances nodulation and growth. Arch Microbiol. https://doi.org/10.1007/s00203-015-1118-z
Trabelsi D, Mhamdi R (2013) Microbial inoculants and their impact on soil microbial communities: a review. Biomed Res Int 2013:863240. https://doi.org/10.1155/2013/863240
Mendes R, Kruijt M, de Bruijn I et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. https://doi.org/10.1126/science.1203980
Défago G, Keel C, Moënne-Loccoz Y (1997) Fate of released Pseudomonas bacteria in the soil profile: implications for the use of genetically modified microbial inoculants. In: Zelikoff JT, Lynch JM, Shepers J (eds) Ecotoxicology: responses, biomarkers and risk assessment. SOS Publications, Fair Heaven, NJ, pp 403–418
Trabelsi D, Mengoni A, Ben Ammar H, Mhamdi R (2011) Effect of on-field inoculation of Phaseolus vulgaris with rhizobia on soil bacterial communities. FEMS Microbiol Ecol 77:211–222. https://doi.org/10.1111/j.1574-6941.2011.01102.x
Schmidt R, Köberl M, Mostafa A et al (2014) Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front Microbiol 5:1–11. https://doi.org/10.3389/fmicb.2014.00064
Andreote FD, da Rocha UN, Araújo WL et al (2010) Effect of bacterial inoculation, plant genotype and developmental stage on root-associated and endophytic bacterial communities in potato (Solanum tuberosum). Antonie Van Leeuwenhoek 97:389–399. https://doi.org/10.1007/s10482-010-9421-9
Gomes NCM, Kosheleva IA, Abraham WR, Smalla K (2005) Effects of the inoculant strain Pseudomonas putida KT2442 (pNF142) and of naphthalene contamination on the soil bacterial community. FEMS Microbiol Ecol 54:21–33. https://doi.org/10.1016/j.femsec.2005.02.005
Özen AI, Ussery DW (2012) Defining the Pseudomonas genus: where do we draw the line with azotobacter? Microb Ecol 63:239–248. https://doi.org/10.1007/s00248-011-9914-8
Loper JE, Hassan KA, Mavrodi DV et al (2012) Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002784
Winsor GL, Lam DKW, Fleming L et al (2011) Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. https://doi.org/10.1093/nar/gkq869
Tkacz A, Poole P (2015) Role of root microbiota in plant productivity. J Exp Bot 66:2167–2175. https://doi.org/10.1093/jxb/erv157
Yang J, Kloepper JW, Ryu C (2008) Rhizosphere bacteria help plants tolerate abiotic stress. Cell Press 14:1–4
D’Souza-Ault MR, Smith LT, Smith GM (1993) Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl Environ Microbiol 59:473–478
Mäkelä P, Jokinen K, Kontturi M et al (1998) Foliar application of glycinebetaine—a novel product from sugar beet—as an approach to increase tomato yield. Ind Crops Prod 7:139–148
Bharti N, Barnawal D, Maji D, Kalra A (2014) Halotolerant PGPRs prevent major shifts in indigenous microbial community structure under salinity stress. Microb Ecol. https://doi.org/10.1007/s00248-014-0557-4
Cheng Z, Woody OZ, McConkey BJ, Glick BR (2012) Combined effects of the plant growth-promoting bacterium Pseudomonas putida UW4 and salinity stress on the Brassica napus proteome. Appl Soil Ecol 61:255–263. https://doi.org/10.1016/j.apsoil.2011.10.006
King AJ, Farrer EC, Suding KN, Schmidt SK (2012) Co-occurrence patterns of plants and soil bacteria in the high-alpine subnival zone track environmental harshness. Front Microbiol 3:1–14. https://doi.org/10.3389/fmicb.2012.00347
Ashraf M, Hasnain S, Berge O, Mahmood T (2004) Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol Fertil Soils 40:157–162. https://doi.org/10.1007/s00374-004-0766-y
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39. https://doi.org/10.1016/j.micres.2013.09.009
Singh A (2015) Soil salinization and waterlogging: a threat to environment and agricultural sustainability. Ecol Indic 57:128–130. https://doi.org/10.1016/j.ecolind.2015.04.027
Ghassemi F, Jakeman AJ, Nix HA (1995) Salinisation of land and water resources: human causes, extent, management and case studies. CAB International, Wallingford
El-Ashry MT, Duda AM (1999) Future perspectives on agricultural drainage. In: Skaggs RW, Van Schilfgaarde J (eds) Agricultural drainage, Agronomy S. American Society of Agronomy, Madison
Fu Q, Liu C, Ding N et al (2010) Ameliorative effects of inoculation with the plant growth-promoting rhizobacterium Pseudomonas sp. DW1 on growth of eggplant (Solanum melongena L.) seedlings under salt stress. Agric Water Manag 97:1994–2000. https://doi.org/10.1016/j.agwat.2010.02.003
Setia R, Gottschalk P, Smith P et al (2012) Soil salinity decreases global soil organic carbon stocks. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2012.08.028
Bruning B, Rozema J (2013) Symbiotic nitrogen fixation in legumes: perspectives for saline agriculture. Environ Exp Bot 92:134–143. https://doi.org/10.1016/j.envexpbot.2012.09.001
Mavi MS, Marschner P (2013) Salinity affects the response of soil microbial activity and biomass to addition of carbon and nitrogen. Soil Res 51:68–75. https://doi.org/10.1071/SR12191
Canfora L, Bacci G, Pinzari F et al (2014) Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE. https://doi.org/10.1371/journal.pone.0106662
Egamberdieva D (2011) Pseudomonas chlororaphis: a salt-tolerant bacterial inoculant for plant growth stimulation under saline soil conditions. Acta Physiol Plant 34:751–756. https://doi.org/10.1007/s11738-011-0875-9
Avila LA (2012) Diversity and biotechnological potential of Pseudomonas spp. from mangrove sediments. Dissertation, University of São Paulo
Vasconcellos RLF, Santos SN, Zucchi TD et al (2017) Pseudomonas aestus sp. nov., a plant growth-promoting bacterium isolated from mangrove sediments. Arch Microbiol. https://doi.org/10.1007/s00203-017-1410-1
Vasconcellos R, Mendes R, Taketani R et al (2013) Draft genome sequence of Pseudomonas sp. strain CMAA 1215, a plant growth-promoting bacterium isolated from a Brazilian mangrove. Genome Announc 1:e00995-e1013. https://doi.org/10.1128/genomeA.00763-13.4
Embrapa (1997) Manual de métodos de análise de solo, 2nd ed. Empresa Brasileira de Pesquisa Agropecuária, Rio de Janeiro
Nadeem SM, Zahir ZA, Naveed M, Arshad M (2007) Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can J Microbiol 53:1141–1149. https://doi.org/10.1139/W07-081
Wang Y, Qian P (2009) Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4:e7401. https://doi.org/10.1371/journal.pone.0007401
Bessieres M-A, Gibon Y, Lefeuvre JC, Larher F (1999) A single-step purification for glycine betaine determination in plant extracts by isocratic HPLC. J Agric Food Chem 47:3718–3722. https://doi.org/10.1021/jf990031h
Rognes T, Flouri T, Nichols B et al (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 2016:1–22. https://doi.org/10.7717/peerj.2584
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303.QIIME
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. https://doi.org/10.1093/nar/gks1219
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. https://doi.org/10.1371/journal.pcbi.1003531
Oksanen J, Blanchet FG, Friendly M et al (2019) vegan: community ecology package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
Mendiburu F (2020) agricolae: statistical procedures for agricultural research. R package version 1.3-3. https://CRAN.R-project.org/package=agricolae
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2:18–22
Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59:206–216. https://doi.org/10.1016/j.envexpbot.2005.12.006
Cunha S, d’Avó AF, Mingote A et al (2013) Mannosylglucosylglycerate biosynthesis in the deep-branching phylum planctomycetes: characterization of the uncommon enzymes from Rhodopirellula baltica. Sci Rep 3:2378. https://doi.org/10.1038/srep02378
Sindhu SS, Gupta SK, Dadarwal KR (1999) Antagonistic effect of Pseudomonas spp. on pathogenic fungi and enhancement of growth of green gram (Vigna radiata). Biol Fertil Soils 29:62–68. https://doi.org/10.1007/s003740050525
Kumawat KC, Sharma P, Sirari A et al (2019) Synergism of Pseudomonas aeruginosa (LSE-2) nodule endophyte with Bradyrhizobium sp. (LSBR-3) for improving plant growth, nutrient acquisition and soil health in soybean. World J Microbiol Biotechnol 35:47. https://doi.org/10.1007/s11274-019-2622-0
Egamberdieva D, Wirth S, Jabborova D et al (2017) Coordination between Bradyrhizobium and Pseudomonas alleviates salt stress in soybean through altering root system architecture. J Plant Interact 12:100–107. https://doi.org/10.1080/17429145.2017.1294212
Kloepper JW, Ryu C-M, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Nassal D, Spohn M, Eltlbany N et al (2018) Effects of phosphorus-mobilizing bacteria on tomato growth and soil microbial activity. Plant Soil 427:17–37. https://doi.org/10.1007/s11104-017-3528-y
Ahmad M, Zahir ZA, Nazli F et al (2013) Effectiveness of halo-tolerant, auxin producing Pseudomonas and Rhizobium strains to improve osmotic stress tolerance in mung bean (Vigna radiata L.). Braz J Microbiol 44:1341–1348
Barea J-M, Pozo MJ, Azcón R, Azcón-Aguilar C (2005) Microbial co-operation in the rhizosphere. J Exp Bot 56:1761–1778. https://doi.org/10.1093/jxb/eri197
Kozdrój J, Trevors JT, van Elsas JD (2004) Influence of introduced potential biocontrol agents on maize seedling growth and bacterial community structure in the rhizosphere. Soil Biol Biochem 36:1775–1784. https://doi.org/10.1016/j.soilbio.2004.04.034
Dedysh SN (2019) Bryobacter. Bergey’s manual of systematics of archaea and bacteria. Wiley, New Jersey, pp 1–5
Kulichevskaya IS, Suzina NE, Liesack W, Dedysh SN (2010) Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemo-organotroph from subdivision 3 of the Acidobacteria. Int J Syst Evol Microbiol 60:301–306. https://doi.org/10.1099/ijs.0.013250-0
Glick BR, Todorovic B, Czarny J et al (2007) Promotion of plant growth by bacterial ACC deaminase. CRC Crit Rev Plant Sci 26:227–242. https://doi.org/10.1080/07352680701572966
Bruce T, Martinez IB, Neto OM et al (2010) Bacterial community diversity in the Brazilian Atlantic forest soils. Microb Ecol 60:840–849. https://doi.org/10.1007/s00248-010-9750-2
Hedlund BP (2010) Phylum XXIII. Verrucomicrobia phyl. nov. Bergey’s manual® of systematic bacteriology. Springer New York, New York, pp 795–841
Brewer TE, Handley KM, Carini P et al (2017) Genome reduction in an abundant and ubiquitous soil bacterium ‘Candidatus Udaeobacter copiosus’. Nat Microbiol 2:16198. https://doi.org/10.1038/nmicrobiol.2016.198
Ofek M, Hadar Y, Minz D (2012) Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS ONE 7:e40117. https://doi.org/10.1371/journal.pone.0040117
Wang X, Sharp CE, Jones GM et al (2015) Stable-isotope probing identifies uncultured planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl Environ Microbiol 81:4607–4615. https://doi.org/10.1128/AEM.00055-15
Dedysh SN (2011) Cultivating uncultured bacteria from Northern Wetlands: knowledge gained and remaining gaps. Front Microbiol. https://doi.org/10.3389/fmicb.2011.00184
García-Salamanca A, Molina-Henares MA, van Dillewijn P et al (2013) Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb Biotechnol 6:36–44. https://doi.org/10.1111/j.1751-7915.2012.00358.x
Acknowledgements
This study was supported by the São Paulo Research Foundation—FAPESP (Grant No. 2012/16623-4) and the Brazilian Agricultural Research Corporation—Embrapa (Unit: Embrapa Environment).
Author information
Authors and Affiliations
Contributions
RLFV designed, directed the project, and wrote the manuscript in consultation with TDZ and ISM. RLFV and ISM conceived of the presented idea. EMR and SNS carried out the metagenome and the greenhouse experiments. RGT contributed to the analysis of the results. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors hereby declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vasconcellos, R.L.F., Romagnoli, E.M., Taketani, R.G. et al. Impact of Inoculation with Pseudomonas aestus CMAA 1215T on the Non-target Resident Bacterial Community in a Saline Rhizosphere Soil. Curr Microbiol 78, 218–228 (2021). https://doi.org/10.1007/s00284-020-02285-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02285-9