Abstract
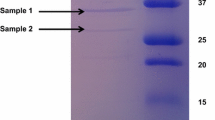
Bacillus paralicheniformis MKU3 produces commercially important keratinolytic proteases by utilizing chicken feather. To unravel the genetics of these degrading keratinolytic proteases in B. paralicheniformis MKU3, we sequenced the genome of this bacterium and studied the protease distribution and their characteristics using bioinformatics tools. Also, a proteomic analysis was performed to identify the consortium of proteases involved in feather hydrolysis. A total of 2,531,755 quality reads were obtained in whole genome sequencing with an approximate coverage fold of 105. The draft genome consists of 4,370,039 bp with 45 contigs. The draft genome codes for 4874 protein-coding genes. Furthermore, 109 genes coding for RNA, including 26 rRNA and 83 tRNA, were identified. Phylogenetic analysis of B. paralicheniformis MKU3 showed closest homolog to B. paralicheniformis F47. Genes coding for proteases belonging to five families were identified with the following proportions 37%, 36%, 9%, 14%, 2%, and 2% of serine-, metallo-, cysteine-, mixed-, and uncharacterized proteases, respectively. Metallo- and serine-protease represented more than 70% of the total proteases. Major protease families distributed in the genome were S8, S9, S33, M20, M50, C26, and C40. Most of the proteases showed significant similarity with the conserved domain database and also identified conserved catalytic sites and domains. SDS-PAGE and zymogram analysis of concentrated feather hydrolysis revealed the active proteases ranging from 10 to 250 kD in size. Proteomic analysis on the feather hydrolysis of B. paralicheniformis MKU3 identified two proteases belonging to serine proteases (S8) and other two as metalloproteases.





Similar content being viewed by others
References
Anantharaman V, Aravind L (2003) Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol 4:R11
Armenteros JJA, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnol 37:420–423
Barret AJ, Rawlings ND, Woessner JF (2004) Hand book of proteolytic enzymes. Introduction: metallopeptidases and their clans volume 1. Second edition Academic press, Elsevier, pp 231–267
Bhunia B, Basak B, Dey A (2012) A review on production of serine alkaline protease by Bacillus spp. J Biochem Tech 3:448–457
Brocklehurst K, Philpott MP (2013) Cysteine proteases: mode of action and role in epidermal differentiation. Cell Tissue Res 351:237–244
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J (2009) DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120
Chen X, Wu J, Chen X (2011) Extracellular metalloproteases from bacteria. Appl Microbiol Biotechnol 92:253–256
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTS. Genome Res 14:1147–1159
Choi N-S, Yoo K, Hahm J, Yoon K, Hang K, Hyun B, Maeng PJ, Kim S (2004) Purification and characteristics of new peptidase, Bacillopeptidase DJ-2, having fibrinolytic activity produced by Bacillus species DJ-2 from Deong Jang. J Microbiol Biotechnol 15:72–79
Contesini FJ, Melo RR, Sato HH (2017) An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol 38(3):321–334
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
Du Y, Ma J, Yin Z, Liu K, Yao G, Xu W, Fan L, Du B, Ding Y, Wang C (2019) Comparative genomic analysis of Bacillus paralicheniformis MDJK30 with its closely related species reveals an evolutionary relationship between B. paralicheniformis and B. licheniformis. BMC Genomics 20:1–16
Dunlap CA, Kwon S, Rooney AP, Kim S (2019) Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. Int J Syst Evol Microbiol 65:3487–3492
Frankowski H, Gu Y, Heo JH, Milner R, Zoppo GJ (2012) Use of gel zymography to examine matrix metalloproteinase (gelatinase) expression in brain tissue or in primary glial cultures. Methods Mol Biol 814:221–233
Gupta R, Ramnani P (2006) Microbial keratinases and their prospective applications: An overview. Appl Microbiol Biotechnol 70:21–33
Huang Y, Busk PK, Lange L (2014) Production and characterization of keratinolytic proteases produced by Onygena corvine. Fungal Genom Biol 5:1–7
Krogh A, Larsson B, Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Lagesen K, Hallin P, Rødland EA, Stærfeldt H, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
Lange L, Huang Y, Busk PK (2016) Microbial decomposition of keratin in nature - a new hypothesis of industrial relevance. Appl Microbiol Biotechnol 100:2083–2096
Li S, Norioka S, Sakiyama F (1998) Bacteriolytic activity and specificity of Achromobacter β-lytic protease. J Biochem 339:332–339
Lowe TM, Eddy SR (1997) tRNAscan-SE : a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226
Mostertz J, Hochgräfe F, Jürgen B, Schweder T, Hecker M (2008) The role of thioredoxin TrxA in Bacillus subtilis: a proteomics and transcriptomics approach. Proteomics 8:2676–2690
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:206–214
Parrado J, Rodriguez-morgado B, Tejada M, Hernandez T, Garcia C (2014) Proteomic analysis of enzyme production by Bacillus licheniformis using different feather wastes as the sole fermentation media. Enzyme Microb Technol 57:1–7
Radha S, Gunasekaran P (2009) Purification and characterization of keratinase from recombinant Pichia and Bacillus strains. Protein Expr Purif 64:24–31
Rao MB, Tanksale AM, Ghatge MS (1998) Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635
Rawlings ND, Barrett AJ (1993) Evolutionary families of peptidases. Biochem J 290:205–218
Rawlings ND, Barrett AJ, Bateman A (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucl Acid Res 40:343–350
Razzaq A, Shamsi S, Ali A, Ali Q, Sajjad M, Malik A, Ashraf M (2019) Microbial proteases applications. Front Bioeng. Biotech 7:1–20
Santha Kalaikumari S, Vennila T, Monika V, Chandraraj K, Rajendhran J (2019) Bioutilization of poultry feather for keratinase production and its application in leather industry. J Clean Prod 208:44–53
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17
Switzer RC, Merril CR, Shifrin S (1979) A highly sensitive silver stain for detecting proteins and peptides in polyacrylamide gels. Anal Biochem 98:231–237
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6. Mol Biol Evol 30:2725–2729
Voigt B, Schweder T, Sibbald MJJB, Albrecht D (2006) The extracellular proteome of Bacillus licheniformis grown in different media and under different nutrient starvation conditions. Proteomics 6:268–281
Wang K, Wang K, Zhou N, Tian Y (2016) Secretory expression, purification, characterization, and application of an Aspergillus oryzae prolyl aminopeptidase in Bacillus subtilis. Appl Biochem Biotechnol 181:1611–1623
Wu J, Chen X (2011) Extracellular metalloproteases from bacteria. Appl Microbiol Biotechnol 92:253–262
Zhao HL, Chen XL, Xie BB, Zhou M, Gao X, Zhang X, Zhou B, Weiss AS, Zhang Y (2012) Elastolytic mechanism of a novel M23 metalloprotease pseudoalterin from deep-sea Pseudoalteromonas sp. CF6-2. J Biol Chem 47:39710–39720
Acknowledgements
UGC-NRCBS, DBT-IPLS, DST-PURSE programs of the School of Biological Sciences, MKU are gratefully acknowledged for providing the facility for genome sequencing and analysis. Authors thank Dr. U.S. Vishnu and Dr. J. Sankarasubramanian for their technical support. We also thank the Mass spectrometry facility of CCAMP/NCBS-TIFR.
Funding
This work was supported by research grant from the Department of Biotechnology, New Delhi, India (No. BT/PR4799/PID/6/642/2012).
Author information
Authors and Affiliations
Contributions
SS performed the experiment, analyzed the data and drafted the manuscript. RS participated in editing the manuscript. JR designed, coordinated, supervised and reviewed the manuscript. PG also reviewed the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors in this article declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2020_2271_MOESM2_ESM.xlsx
List of proteases identified from B. paralicheniformis strains. Ten strains of B. paralicheniformis have been retrieved in this study and were analyzed for protease distribution.
284_2020_2271_MOESM3_ESM.xlsx
Extracellular proteases and proteins from B. paralicheniformis MKU3 listed in this file. These proteases and proteins were induced by chicken feather degradation by B. paralicheniformisMKU3.
Rights and permissions
About this article
Cite this article
SanthaKalaikumari, S., Sivakumar, R., Gunasekaran, P. et al. Whole-genome Sequencing and Mining of Protease Coding Genes in Bacillus paralicheniformis MKU3, and its Degradomics in Feather Meal Medium. Curr Microbiol 78, 206–217 (2021). https://doi.org/10.1007/s00284-020-02271-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02271-1