Abstract
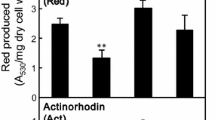
Activating the genetic potential of Streptomyces strains to produce secondary metabolites can improve the production of useful biologically active compounds and facilitate the discovery of novel biologically active compounds. In this study, we found that Streptomyces lividans carrying the R440H mutation in rpoB, encoding the RNA polymerase beta subunit, grown in the presence of lincomycin at concentrations below the minimum inhibitory concentration (MIC) produced abundant amounts of actinorhodin and certain cryptic secondary metabolites despite culture conditions that restrict their production by the wild-type strain. The results indicate that lincomycin at concentrations below the MIC may strongly potentiate secondary metabolite production by Streptomyces strains carrying a specific rpoB mutation. In this study, we report an interesting phenomenon induced by combining the positive effects of certain rpoB mutations and concentration-dependent responses to lincomycin on secondary metabolism in S. lividans 66 and discuss the mechanisms and their applicability in exploring cryptic secondary metabolite production in streptomycetes.




Similar content being viewed by others
References
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich
Kemung HM, Tan LT, Khan TM, Chan KG, Pusparajah P, Goh BH, Lee LH (2018) Streptomyces as a prominent resource of future anti-MRSA drugs. Front Microbiol. https://doi.org/10.3389/fmicb.2018.02221
Bentley S, Chater K, Cerdeño-Tárraga AM, Challis G, Thomson N, James K, Harris D, Quail M, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen C, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell B, Parkhill J, Hopwood D (2002) Completegenome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Ward AC, Nee Allenby (2018) Genome mining for the search and discovery of bioactive compounds: the Streptomyces paradigm. FEMS Microbiol Lett. https://doi.org/10.1093/fensle/fny240
Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 42:343–370
Onaka H (2017) Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J Antibiotechnol 70:865–870
Tomm HA, Ucciferri L, Ross AC (2019) Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J Ind Microbiol Biotechnol 46:1381–1400
Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K (2004) Ribosome engineering and secondary metabolite production. Adv Appl Microbiol 56:155–184
Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K (2009) Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27:462–464
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97:87–98
Imai Y, Sato S, Tanaka Y, Ochi K, Hosaka T (2015) Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl Environ Microbiol 81:3869–3879
Ishizuka M, Imai Y, Mukai K, Shimono K, Hamauzu R, Ochi K, Hosaka T (2018) A possible mechanism for lincomycin induction of secondary metabolism in Streptomyces coelicolor A3 (2). Antonie Van Leeuwenhoek 111:705–716
Hoshino K, Imai Y, Mukai K, Hamauzu R, Ochi K, Hosaka T (2020) A putative mechanism underlying secondary metabolite overproduction by Streptomyces strains with a 23S rRNA mutation conferring erythromycin resistance. Appl Microbiol Biotechnol 104:2193–2203
Mak S, Nodwell JR (2017) Actinorhodin is a redox-active antibiotic with a complex mode of action against Gram-positive cells. Mol Microbiol 106:597–613
Lin L, Pan J, Wang Z, Yan X, Yang D, Zhu X, Shen B, Duan Y, Huang Y (2018) Ribosome engineering and fermentation optimization leads to overproduction of tiancimycin A, a new enediyne natural product from Streptomyces sp. CB03234. J Ind Microbiol Biotechnol 45:141–151
Kaweewan I, Komaki H, Hemmi H, Hoshino K, Hosaka T, Isokawa G, Oyoshi T, Kodani S (2019) Isolation and structure determination of a new cytotoxic peptide curacozole from Streptomyces curacoi based on genome mining. J Antibiot 72:1–7
Hu H, Zhang Q, Ochi K (2002) Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J Bacteriol 184:3984–3991
Lai C, Xu J, Tozawa Y, Okamoto-Hosoya Y, Yao X, Ochi K (2002) Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology 148:3365–3373
Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K (2002) A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol Genet Genomics 268:179–189
Baltz RH (2014) Spontaneous and induced mutations to rifampicin, streptomycin and spectinomycin resistances in actinomycetes: mutagenic mechanisms and applications for strain improvement. J Antibiotechnol 67:619–624
Ma Z, Luo S, Xu X, Bechthold A, Yu X (2016) Characterization of representative rpoB gene mutations leading to a significant change in toyocamycin production of Streptomyces diastatochromogenes 1628. J Ind Microbiol Biotechnol 43:463–471
Acknowledgments and Funding Information
This work was financially supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (Grant No. 18K05410) to T. H.
Author information
Authors and Affiliations
Contributions
KM, MK, DH and TH designed research; KM, MK, KH, RH and TH performed research; KM, MK, and TH analyzed data: KM, TM and TH wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research Involving Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mukai, K., Kobayashi, M., Hoshino, K. et al. Lincomycin-Induced Secondary Metabolism in Streptomyces lividans 66 with a Mutation in the Gene Encoding the RNA Polymerase Beta Subunit. Curr Microbiol 77, 2933–2939 (2020). https://doi.org/10.1007/s00284-020-02126-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02126-9