Abstract
Surface display using spores of Bacillus subtilis is widely used to anchor antigens and enzymes of different sources. One open question is whether anchored proteins are able to form disulfide bonds. To answer this important question, we anchored the Escherichia coli alkaline phosphatase PhoA on the spore surface using two different surface proteins, CotB and CotZ. This enzyme needs two disulfide bonds to become active. Subsequently, we purified the spores and assayed for alkaline phosphatase activity. In both cases, we were able to recover enzymatic activity. Next, we asked whether formation of disulfide bonds occurs spontaneous or is catalyzed by thiol-disulfide oxidoreductases upon lysis of the cells. The experiment was repeated in a double-knockout mutant ΔbdbC and ΔbdbD. Since the disulfide bonds are also present on spores prepared from the double knockout, we conclude that oxidative environment after cell lysis is sufficient for disulfide formation of alkaline phosphatase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Surface display is a molecular technique to present heterologous proteins or peptides on the surface of bio-particles, such as phages, bacterial cells, yeast, and higher eukaryotic cells or endospores. Proteins of interest (often called passenger proteins) are fused to naturally occurring surface proteins of these particles, e.g., coat proteins. When the fusion proteins are synthesized, the passenger proteins will be exposed on the surface of the bio-particles. Surface display was invented by Smith [22]. Since then, surface display has been used for a multitude of applications including affinity screening of peptide libraries to identify ligands for peptide receptors, isolation of monoclonal antibodies, and identification of new antibiotics, as well as high throughput screening for binding partners, production of active enzymes for cleanup of industrial and environmental pollution, development of biosensors and biocatalysts, and even the delivery of vaccines and drugs [2, 15, 21].
Developing a surface display system using endospores of B. subtilis (called spore display) has excessive advantages particularly in the field of vaccine development. B. subtilis is non-pathogenic and non-toxicogenic making oral ingestion possible. Antigen-specific immune responses could be triggered in mice immunized with B. subtilis spores displaying heterologous antigens on their surface [24]. Furthermore, the exceptional longevity of the spore in the environment and its natural resistance to chemicals and heat eases storing and makes refrigeration obsolete [4, 19].
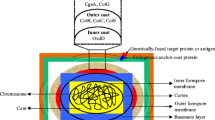
To anchor the proteins of interest on the endospore, spore coat proteins are used as the carrier proteins. Although the spore coat composed more than 70 different proteins, most studies in the last decade successfully displaying various peptides or proteins involved the use of only three coat proteins: CotB, CotC, or CotG [4, 10, 15, 20]. Recent studies suggested a new anchor protein useful in spore surface display, CotZ, which is another member of the outermost layer of the spore coat. This protein was proven to allow efficient display of relatively large passenger proteins on the spore surface [10, 12].
Disulfide bonds are intra- or intermolecular bridges that link the thiol groups of two cysteine residues after thiol oxidation. B. subtilis encodes three membrane-bound (BdbA, BdbB, and BdbC) and one secreted (BdbD) thiol-disulfide oxidoreductase/isomerase enzymes involved in disulfide bond formation. The BdbD protein serves as the major thiol oxidase and is kept in its oxidized state by the paralogous quinone reductases BdbB and BdbC, where the latter enzyme plays a major role [16, 18].
The disulfide-containing protein PhoA of E. coli was used to study disulfide bond formation of secreted heterologous proteins by B. subtilis. PhoA, the alkaline phosphatase of E. coli, contains two disulfide bonds that are indispensable for both the enzymatic activity and the stability of the protein [23]. The contribution of BdbB, BdbC, and BdbD to the folding of exported PhoA into a stable and active conformation could be verified, when bdbB, bdbC, or bdbD mutant strains secreted significantly lower amounts of active PhoA than the parental strain 168. The protein was synthesized and translocated through the membrane, but not correctly folded and thus quickly degraded. Most likely, this lack of folding would be caused by ineffective disulfide bond formation due to the absence of the Bdb proteins [3, 5, 17, 18]. Here, we asked whether PhoA anchored on the spore surface will be present in its active form, and, if yes, whether formation of the two disulfide bonds occurs spontaneously or is catalyzed by the thiol-disulfide oxidoreductases.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Escherichia coli strain JM109 [25] was used as recipient in all cloning experiments. The two-fold protease-deficient strain B. subtilis DB104 [14] (nprE18 ΔaprA3) served as host strain for surface display of the alkaline phosphatase on Bacillus spores. Strain bdbCD [16] (trpC bdbCD::Sp R), which lacks the thiol-disulfide oxidoreductases BdbC and BdbD, was used to test for the influences of these enzymes on the formation of disulfide bonds of the alkaline phosphatase displayed on spores. The plasmids pAG04 [13] and pKH128 [10] were used as cloning vectors. Cells were routinely grown aerobically in Luria–Bertani (LB) medium (1 % tryptone, 0.5 % yeast extract, 1 % NaCl) at 37 °C. To prepare spores, B. subtilis cells were grown in sporulation medium 2 × SG (1,6 % Difco Nutrient Broth, 0.2 % KCl, and 0.05 % MgSO4 × 7 H2O, pH 7, supplemented with 0.1 % glucose, 1 mM Ca(NO3), 0.1 mM MnCl2, and 0.01 mM FeSO4). Antibiotics were added as appropriate (ampicillin at 50 µg/ml; chloramphenicol at 10 µg/ml; erythromycin at 1 or 100 µg/ml; spectinomycin at 1 µg/ml).
Construction of Strains and Plasmids
The bdbCD mutations were transferred from strain bdbCD into strain DB104. To accomplish this goal, chromosomal DNA was isolated from strain bdbCD and transformed into strain DB104, and transformants were selected on LB plates containing spectinomycin. Correct replacement of the bdbCD genes by the mutation was verified by PCR. For displaying recombinant proteins on the spore surface, the coat proteins CotB and CotZ were chosen as carrier proteins. Plasmid pAG04 carries part of the coding region of cotB followed by the coding region for the flexible linker GGGEAAAKGGG and allows the addition of eight histidine residues to the C-terminal end of the fusion protein. Plasmid pKH128 carries the coding region of cotZ followed by the coding region of the same flexible linker and the coding region for the His-tag. Fusions to both cot genes can be inserted at the amyE locus of the B. subtilis chromosome. The DNA of the E. coli phoA gene was amplified by PCR using chromosomal DNA and the primer pair PhoA5 (GGCCAT GGATCC AAACAAAGCACTATTGCACTGGC; BamHI site underlined) and PhoA3 (GGCCAT GAGCTC TTAATGATGATGATGAT GATGATGATGTTTCAGCCCCAGAGCGGCTTTCATG; SacI site underlined). The PCR products were cut with BamHI and SacI and ligated separately into the plasmids pAG04 and pKH128 treated with the same enzymes. Correct insertion of both genes into the plasmids was verified by PCR. The two new recombinant plasmids were designated pAG04-phoA and pKH128-phoA. Next, both recombinant plasmids were linearized with PstI and transformed into strain DB104 followed by selection on LB plates containing chloramphenicol. To identify transformants where the gene fusions have been inserted at the amyE locus, 25 colonies of each were analyzed by spreading them onto LB plates containing 1 % (w/v) soluble starch. After incubation overnight, the cells were flooded with 5 ml of Gram’s iodine stain (0.5 % w/v iodine, 1 % w/v potassium iodide) [8]. After 5 min, the overlaying fluid was discarded, and the colorization was documented immediately.
Purification of Spores
50 ml Schaeffer’s medium with 10 µg/ml chloramphenicol was inoculated with 500 µl of the respective stock. For the control with DB104, no antibiotic was added. The mother cells were sampled at 48 h after inoculation at 37 °C. The OD600 was measured, and 1 ml was taken for phosphatase activity assay with mother cells. The spores of the remaining culture were then isolated. The culture was centrifuged for 10 min at 10,000 rpm, and then the pellet was washed with 50 ml of distilled water and centrifuged again under the same conditions. The pellet was resuspended with 200 µl 20 % (w/v) sodium diatrizoate hydrate and layered onto 1 ml 50 % (w/v) sodium diatrizoate hydrate. This was centrifuged for 15 min at 10,000 rpm and then the supernatant was carefully removed. The pellet was resuspended in water, transferred into a new tube, and washed three times with 1 ml water.
Conformation of Fusion Protein Production by Flow Cytometry
Isolated spores were washed with 1 ml PBS and then resuspended in 200 µl fluorescein isothiocyanate labeled Rabbit Poly Anti-6-His antibodies (KO212220, Komabiotech, Seoul) in PBS. After 30 min on ice, spores were washed three times with 1 ml PBS and resuspended in 200 µl PBS. In a FACS reaction tube, 50 µl spores were mixed with water and analyzed for fluorescence in a flow cytometer.
Phosphatase Activity Assay
Samples of mother cells or isolated spores were washed with 1 ml assay buffer, and the OD600 was measured. The samples were diluted with assay buffer (1 M Tris–HCl, pH 8.0, 10 mM MgSO4, 1 mM ZnCl2, 1 mM iodoacetamide) to OD600 = 1 for mother cell samples and OD600 = 20 for spores. 1 ml of these referenced samples was mixed with 100 µl para-nitrophenyl phosphate (pNPP) solution and incubated for 10 min at 37 °C. To stop the reaction, 120 µl of 0.5 M EDTA, pH 8.0, containing 1 M KH2PO4 was added. Samples were then centrifuged for 2 min at 12,000 rpm, and the OD410 of the supernatant was determined. The activity was calculated by the following equation: Units of activity = Ab410*352/(time(min)*OD600) [8].
Results
Construction of Two B. subtilis Strains Able to Produce Spores Displaying Alkaline Phosphatase
To allow analysis of disulfide bond formation of proteins displayed on the B. subtilis spore surface, we constructed two different strains. In one strain, we fused the coding region of phoA to the C-terminal end of cotB and in the second to that of cotZ. The coding region of a linker peptide was inserted between cotB or cotZ, respectively, and phoA to allow independent folding of both proteins (see Fig. 1). Furthermore, the coding region for a His-tag (His8) was co-translationally fused to the C-terminus of phoA to allow determination of the amount of fusion protein produced by flow cytometry using antibodies recognizing and binding to the His-tag. We fused PhoA to CotB using the vector pKL128 and to CotZ using pAG04. In both plasmid vectors, the fusion proteins were sandwiched between amyE front and amyE back. Next, the recombinant plasmids, which are unable to replicate in B. subtilis, were transformed into B. subtilis strain DB104 selecting for colonies able to grow on plates containing chloramphenicol. Insertion of cotB-phoA or cotZ-phoA at the amyE locus of DB104 was verified by PCR and confirmed by the loss of α-amylase production of DB104 cotB-phoA or DB104 cotZ-phoA on starch plates.
Confirmation of Fusion Protein Production and Sortase Localization by Flow Cytometry
Next, we analyzed whether the fusion proteins are indeed synthesized and anchored on the spore surface. Isolated spores were incubated with fluorescently labeled antibodies recognizing the His-tag. The labeled spores were applied to a flow cytometer, where single spores can be analyzed for fluorescence signals. As shown in Fig. 2, the intensity of the signal is plotted against the counts (number of spores with this intensity) of spores for both fusion strains and strain DB104 as control. As compared to strain DB104 (Fig. 2a), both CotZ-PhoA (Fig. 2b) and CotB-PhoA (Fig. 2c) exhibited an increased intensity, but the amount of displayed and accessible fusion protein varied between the two fusions. CotB-PhoA showed a larger intensity shift than CotZ-PhoA. This might be due to a higher number of CotB protein molecules anchored on the spore surface as compared to CotZ.
Measurements of Alkaline Phosphatase Activities
After we have proven that the alkaline phosphatase is anchored on the outside of the spores, we analyzed them for phosphatase activity. The enzymatic activity was measured by incubating purified spores with the chromogenic substrate p-nitrophenyl phosphate as described under Materials and Methods. The product of this reaction is the yellow p-nitrophenol, which is directly proportional to the amount of active phosphatase and the incubation time. Figure 3 shows the phosphatase activity measured with whole cells and isolated spores from the wild-type strain DB104 containing cotB inserted at the amyE locus (control) and the same strain harboring cotB-phoA or cotZ-phoA. The three types of cells expressed high phosphatase activities. At least four genes of B. subtilis code for alkaline phosphatases, where two of them, phoA and phoB, encode enzymes responsible for 98 % of the cellular alkaline phosphatase activity synthesized during phosphate depletion and developmental control during certain stages of sporulation independent of the phosphate level [11]. The latter conditions are present here and most probably account for the high activities measured with whole cells and did not vary considerably among the three strains (Fig. 3). When the phosphatase activity was measured with the spores prepared from these strains, that from the control spores were rather low, while the activity from spores displaying CotB-PhoA and CotZ-PhoA was significantly higher. The activity of CotZ-PhoA spores was determined to be about 0.3 units and that of CotB-PhoA spores about 0.6 units most probably reflecting the higher amounts of hybrid proteins molecules displayed on the CotB-PhoA spores as mentioned before.
We conclude from these results that both DB104 CotB-PhoA and DB104 CotZ-PhoA spores display active alkaline phosphatase. That means that the correct disulfide bonds have been formed. Who is responsible for formation of the disulfide bonds? We envisage two possibilities. First, the disulfide bonds are formed spontaneously upon lysis of the mother cells due to access of oxygen to the spores. Second, the oxidoreductases anchored in the cytoplasmic membrane and facing the outside of the cells have access to the spores upon cell lysis. To distinguish between these possibilities, we repeated the experiment in a strain unable to synthesize the two major oxidoreductases.
Analysis of the Phosphatase Activity on Spores Prepared from a B. subtilis Mutant Strain Unable to Produce Thiol-Disulfide Oxidoreductases
We introduced first the ΔbdbCD mutation of strain ΔbdbCD [16] into strain DB104 resulting in DB104 ΔbdbCD followed by insertion of cotB-phoA yielding DB104 ΔbdbCD cotB-phoA. Flow cytometry analysis of strain ΔbdbCD cotB-phoA revealed that spores purified from this strain displayed CotB-PhoA on their surface albeit at a reduced amount compared to strain DB104 cotB-phoA (data not shown). Next, we measured the alkaline phosphatase activity with spores prepared from all four strains as shown in Fig. 4. Again, as noted before, the mother cells exhibited a higher phosphatase activity compared to the spores prepared from them (Fig. 4). Most importantly, the alkaline phosphatase activities measured with spores prepared from wild-type and ΔbdbCD strains did not vary significantly. We conclude from this result that the oxidative conditions after cell lysis rather then the oxidoreductases are responsible for formation of the disulfide bonds.
Discussion
Some proteins need further stabilization after folding by the formation of one or more disulfide bonds. This has been first described by Anfinsen and coworkers. They studied refolding of denatured bovine pancreatic ribonuclease A (RNase A). This enzyme contains four disulfide bonds, which are all essential for full activity. After denaturation, RNase A could fold spontaneously to its active oxidized form in the presence of molecular oxygen [1]. While these data led to the conclusion that disulfide bond formation occurs always spontaneously, also in living cells, Anfinsen and coworkers postulated an enzyme correcting wrongly formed disulfide bonds leading to the discovery of the eukaryotic protein disulfide isomerase present in the endoplasmatic reticulum [6, 7]. Later, with the discovery of DsbA in E. coli, it became clear that disulfide bond formation catalysts were necessary for efficient formation of disulfide bonds [9]. Subsequently, oxidoreductases have also been described in B. subtilis [18] and many other bacterial species.
These data help to explain why formation of disulfide bonds of PhoA can be formed spontaneously on spores released from lysed B. subtilis mother cells. But we cannot exclude the possibility that minor oxidoreductase activities present in the ΔbdbCD double knockout are involved in the formation of the disulfide bonds. Another open question which remains to be answered is the percentage of PhoA molecules which contain the correct disulfide bonds. In summary, we could show that proteins displayed on the surface of spores can acquire disulfide bonds. This will be of importance when proteins of biotechnological importance will be anchored on spores.
References
Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230
Benhar I (2001) Biotechnological applications of phage and cell display. Biotechnol Adv 19:1–33
Bolhuis A, Venema G, Quax WJ, Bron S, Van Dijl JM (1999) Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J Biol Chem 274:24531–24538
Cutting SM, Hong HA, Baccigalupi L, Ricca E (2009) Oral vaccine delivery by recombinant spore probiotics. Int Rev Immunol 28:487–505
Darmon E, Dorenbos R, Meens J, Freudl R, Antelmann H, Hecker M, Kuipers OP, Bron S, Quax WJ, Dubois JY, Van Dijl JM (2006) A disulfide bond-containing alkaline phosphatase triggers a BdbC-dependent secretion stress response in Bacillus subtilis. Appl Environ Microbiol 72:6876–6885
De LF, Goldberger RF, Steers E Jr, Givol D, Anfinsen B (1966) Purification and properties of an enzyme from beef liver which catalyzes sulfhydryl-disulfide interchange in proteins. J Biol Chem 241:1562–1567
Goldberger RF, Epstein CJ, Anfinsen CB (1963) Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem 238:628–635
Harwood CR, Cutting S (1990) Molecular biological methods for Bacillus. Wiley, Chichester
Hatahet F, Boyd D, Beckwith J (2014) Disulfide bond formation in prokaryotes: history, diversity and design. Biochim Biophys Acta 1844:1402–1414
Hinc K, Iwanicki A, Obuchowski M (2013) New stable anchor protein and peptide linker suitable for successful spore surface display in B. subtilis. Microb Cell Fact 12:22
Hulett FM, Lee J, Shi L, Sun G, Chesnut R, Sharkova E, Duggan MF, Kapp N (1994) Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J Bacteriol 176:1348–1358
Imamura D, Kuwana R, Takamatsu H, Watabe K (2010) Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J Bacteriol 192:518–524
Iwanicki A, Piatek I, Stasilojc M, Grela A, Lega T, Obuchowski M, Hinc K (2014) A system of vectors for Bacillus subtilis spore surface display. Microb Cell Fact 13:30
Kawamura F, Doi RH (1984) Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol 160:442–444
Kim J, Schumann W (2009) Display of proteins on Bacillus subtilis endospores. Cell Mol Life Sci 66:3127–3136
Kouwen TR, van der Goot A, Dorenbos R, Winter T, Antelmann H, Plaisier MC, Quax WJ, Van Dijl JM, Dubois JY (2007) Thiol-disulphide oxidoreductase modules in the low-GC Gram-positive bacteria. Mol Microbiol 64:984–999
Kouwen TR, Van Dijl JM (2009) Applications of thiol-disulfide oxidoreductases for optimized in vivo production of functionally active proteins in Bacillus. Appl Microbiol Biotechnol 85:45–52
Kouwen TR, Van Dijl JM (2009) Interchangeable modules in bacterial thiol-disulfide exchange pathways. Trends Microbiol 17:6–12
McKenney PT, Driks A, Eichenberger P (2013) The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44
Pan JG, Kim EJ, Yun CH (2012) Bacillus spore display. Trends Biotechnol 30:610–612
Samuelson P, Gunneriusson E, Nygren PA, Stahl S (2002) Display of proteins on bacteria. J Biotechnol 96:129–154
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317
Sone M, Kishigami S, Yoshihisa T, Ito K (1997) Roles of disulfide bonds in bacterial alkaline phosphatase. J Biol Chem 272:6174–6178
Tam NK, Uyen NQ, Hong HA, Duc H, Hoa TT, Serra CR, Henriques AO, Cutting SM (2006) The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol 188:2692–2700
Yanish-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Acknowledgments
This work was funded by the PT-DLR project 01DR13001 on the German side and by the National Research Foundation of Korea (NRF-2012K1A31A0403470).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Richter, A., Kim, W., Kim, JH. et al. Disulfide Bonds of Proteins Displayed on Spores of Bacillus subtilis Can Occur Spontaneously. Curr Microbiol 71, 156–161 (2015). https://doi.org/10.1007/s00284-015-0839-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0839-1