Abstract
Biofiltration is a water purification technology playing a pivotal role in producing safe drinking water. This technology attracts many interests worldwide due to its advantages, such as no addition of chemicals, a low energy input, and a high removal efficiency of organic compounds, undesirable taste and odours, and pathogens. The current review describes the microbial ecology of three biofiltration processes that are routinely used in drinking water treatment plants, i.e. (i) rapid sand filtration (RSF), (ii) granular activated carbon filtration (GACF), and (iii) slow sand filtration (SSF). We summarised and compared the characteristics, removal performance, and corresponding (newly revealed) mechanisms of the three biofiltration processes. Specifically, the microbial ecology of the different biofilter processes and the role of microbial communities in removing nutrients, organic compounds, and pathogens were reviewed. Finally, we highlight the limitations and challenges in the study of biofiltration in drinking water production, and propose future perspectives for obtaining a comprehensive understanding of the microbial ecology of biofiltration, which is needed to promote and optimise its further application.
Key points
• Biofilters are composed of complex microbiomes, primarily shaped by water quality.
• Conventional biofilters contribute to address safety challenges in drinking water.
• Studies may underestimate the active/functional role of microbiomes in biofilters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drinking water sources including surface water from rivers, lakes, and reservoirs, as well as groundwater aquifers, may contain a variety of contaminants, such as organic compounds, chemical substances, and pathogenic viruses, bacteria, and protozoa (Pandey et al. 2014; Palansooriya et al. 2020). Due to increasing human activities, the safety of drinking water sources is currently facing enormous challenges worldwide and has been associated with increased risk of contamination (Shannon et al. 2008; World Health Organization 2017; Zhang et al. 2017; Favere et al. 2021). Contamination of drinking water can lead to waterborne diseases (e.g. cholera, typhoid, hepatitis A, and dysentery) and, hence, threatens public health worldwide (Moreira and Bondelind 2017; World Health Organization 2017). To address drinking water-related health problems, processes of establishing sufficient barriers against contaminants in water sources, further referred to as water treatment, is essential prior to consumption.
Biofiltration treatment is now attracting more interest worldwide due to its advantages of avoiding addition of chemicals, low energy input, and higher removal efficiency in turbidity, organic compounds, undesirable tastes and odours, and especially pathogens (e.g. bacteria, viruses, and protozoa), meanwhile meeting the increased demand of safe and high-quality drinking water (Haig et al. 2011; Nyberg et al. 2014; Basu et al. 2015; Zhang et al. 2017; Oh et al. 2018; Maurya et al. 2020). Biofiltration is a process that not only removes fine particles through physicochemical means (e.g. straining and sorption) like other conventional filters but also captures and degrades contaminants through biological activities (Basu et al. 2015; Liu et al. 2017; Terry and Summers 2018). It has been used in Europe for purifying surface water to effectively reduce turbidity and cholera bacteria in drinking water applications since the early 1900s. However, the importance of biofiltration in drinking water treatment was noticed only after it was found to benefit in reducing microbial growth (in the distribution pipelines), corrosion potential, and the disinfection by-products a few decades ago (Chaudhary et al. 2003). With the increasing presence of so-called contaminants of emerging concern (CECs), such as pharmaceutical residues and industrial chemicals in source waters (Zhang et al. 2017; Yusuf et al. 2021), a good understanding of biofiltration processes is essential for further improvement in the production of safe drinking water.
Here, we comprehensively review and compare 3 major biofiltration processes in drinking water treatment plants (DWTPs), including rapid sand filtration (RSF), granular activated carbon filtration (GACF), and slow sand filtration (SSF), from both the performance and mechanistic perspectives. The review specifically focusses on the microbial ecology of biofilters and the role of microbial communities in the removal of nutrients, organic compounds, and pathogens. In addition, the limitations, biases, and challenges of the research to date are discussed. This study proposes directions that will complement current studies to obtain a comprehensive understanding of the microorganisms involved in the production of safe drinking water.
Biofilters in the drinking water treatment process
Since the early twentieth century, multiple-stage water treatments are carried out in the majority of DWTPs. DWPTs comprise 3 main biofiltration processes including RSF, GACF, and SSF, which are applied together with coagulation, flocculation, sedimentation, and disinfection steps in various combinations (Fig. 1a) (Basu et al. 2015; World Health Organization 2017).
Conceptional model of biofiltration processes, the microorganisms present, and the possible mechanisms in the removal of different contaminants. a Overview of the different biofiltration processes in the production of drinking water. b Microbial community of biofilters. A summary of microorganisms present in biofilters is listed in 3 green boxes (including dominantly occurring bacterial phyla). Potentially functional organisms are described in the right box. The occurrence of potentially functional groups of organisms is indicated by the numbers below each box. Examples of functional groups that exist in the group are indicated by corresponding colours under the phylum. c Contaminants present in the influent water (orange box) grouped into inorganic, organic, and biological contaminants. Biofiltration removal efficiency (green box, left columns): the intensity of the colour represents the relative amount of the contaminant in the effluent water, i.e. the more intense, the more contaminants break through the filters and will be present in the effluent water, and so less removal efficiency; conversely, the lighter the column, less contaminants enter the effluent, and the better removal efficiency;
 highlight the major removal of the biofiltration step. Biofilter removal mechanisms (green box, right columns): the mechanisms involved in the removal of the corresponding contaminants are grouped and presented via
highlight the major removal of the biofiltration step. Biofilter removal mechanisms (green box, right columns): the mechanisms involved in the removal of the corresponding contaminants are grouped and presented via
 Biological removal and/or
Biological removal and/or
 Physical–chemical removal. The numbers marked on the colour block (1–6, explained in b) represent the organism groups involved in the removal process
Physical–chemical removal. The numbers marked on the colour block (1–6, explained in b) represent the organism groups involved in the removal process
Filtration performance of biofilters
Rapid sand filtration
RSF processes water with a wide range of initial turbidity at a high flow rate (5–30 m/h), which can reduce most physical hazards (particles) and inorganic compounds (e.g. iron, manganese, ammonia, nitrate) (World Health Organization 2017; Brandt et al. 2017). In a combination of pre-treatments including coagulation and sedimentation, an appropriate RSF process can reduce the water turbidity to less than 0.1 nephelometric turbidity units (NTU) (World Health Organization 2017). When the water turbidity is reduced to ≤ 0.3 NTU in 95% of the samples and none to exceed 1 NTU, about 1–2 log10 reduction of viruses and 3 log10 reductions of Cryptosporidium are considered to be achieved (World Health Organization 2017; Brandt et al. 2017). Besides, RSF has a proven effect on removing iron (28.6–97%) and manganese (50–97%) and to a less extent on removing unpleasant odour and taste, bacteria (0.3–3 log10), and organic matters (e.g. 99% methane) (Bishaw and Kebede 1999; Lee et al. 2014; Asami et al. 2016; Poghosyan et al. 2020) (Table S1).
Granular activated carbon filtration
GACF is applied to further remove chemical and biological hazards that can break through the RSF and the following ozonation barriers. Usually, water is first treated by ozone after RSF and then by GACF at a relatively fast flow rate (6−7.5 m/h) (World Health Organization 2017; Brandt et al. 2017; de Vera et al. 2019).
GACF is primarily designed to remove unpleasant odour, taste, and colour from water caused by natural organic matter (NOM) and organic micropollutants (Magic-Knezev et al. 2008; Simpson 2008). In the last decades, GACF has been observed to effectively remove a wide range of CECs (e.g. 22 to > 80% of 16 studied CECs including pharmaceutical residues, pesticides, and fire retardant) and assimilable organic carbon (AOC) (39−74%) associated with microbial stability of water (Wang et al. 2013; World Health Organization 2017; Zhang et al. 2017; Greenstein et al. 2018; de Vera et al. 2019; Pick et al. 2019) (Table S3). Regarding microbial contaminants, GACF removes protozoan (oo)cyst (1.0−2.3 log10) such as Cryptosporidium parvum and Giardia lamblia. However, GACF shows limited removal efficiency of faecal indicator bacteria (i.e. ≤ 0.5−1.1 log10 Escherichia coli) and anaerobic spores (0.9−1.1 log10 spores of Clostridium bifermentans), and even lower removal efficiency of viruses and bacteriophages (0−0.7 log10 MS2 phage) (Hijnen et al. 2010; Hijnen and Medema 2010).
Slow sand filtration
Differing from RSF filters, finer sand is the filter medium in SSF. The sand bed filters water with a low flow rate (0.1−0.5 m/h) (Brandt et al. 2017). SSF is effective for reducing turbidity, total organic carbon (TOC), nitrogen compounds, pesticides, pharmaceutical chemicals, and microbial contaminants (Table S5) (Hijnen et al. 2004; Bichai et al. 2010; D'Alessio et al. 2015). SSF is one of the oldest and most effective approaches to control microbial contamination (i.e. pathogenic oocysts, bacteria, and viruses) from drinking water (Hijnen et al. 2007; Haig et al. 2011). SSF typically removes 2−6 log10 oocysts, 2−4 log10 of bacteria, and < 1−3 log10 of viruses (Hijnen et al. 2004; Wakelin et al. 2010; Matuzahroh et al. 2020) (Table S5).
SSF performance depends on source water characteristics, temperature, sand type, grain size, bed depth, filtration rate, and the age and thickness of the “Schmutzdecke” (Matuzahroh et al. 2020; Maurya et al. 2020; Schijven et al. 2013; Yogafanny et al. 2014). The removal efficiency increases along with a finer grain size, a longer bed depth, a slower flow rate, and an older age of Schmutzdecke (Bauer et al. 2011; Schijven et al. 2013; Yogafanny et al. 2014). The Schmutzdecke (SD) is a slimy biofilm layer on top of the slow sand filter with a thickness of 0.5 up to 3 cm, which is described in more detail in the later sections.
Mechanism of biofiltration processes
In biofiltration processes, the biological removal always occurs in parallel with physical–chemical removal (Wu et al. 2014; Verma et al. 2017), although one removal mechanism could outperform the other one (Fig. 2).
Conceptional model of dynamic accumulation of removal mechanisms in RSF, GACF, and SSF filters. The x-axis lists the conditions that could stimulate (positive correlate to) biological removal; The y-axis lists conditions that can increase the physical–chemical removal portion. The white curve depicts the dynamic changes in the proportion of physical–chemical and biological removal mechanisms in the total removal performance. The change in slope (in GACF) refers to the transition from normal GACF to biological-active GACF. *Depending on material characteristics, filter media could positively induce both the physical–chemical and biological removal
When operating a new filter, the removal is initially achieved through the physical mechanisms straining, and adsorption (Hijnen and Medema 2010; Knezev 2015). Meanwhile, suspended particles gradually form a nutrient-rich rough layer on the filter surface, attracting microorganisms to colonise the surface. Biofilms are then generated on the surface over time. From this stage onwards, biological removal (e.g. biodegradation) gradually becomes the main removal mechanism (Hammes et al. 2008; Velten et al. 2011; Brown et al. 2015).
Biological removal depends on the biological activity of the biotic community accumulated in filters. Various operating conditions of the filtration process can affect the biotic community, such as filter media, nutrients from influent water, flow rate, temperature, and maintenance measures (i.e. backwashing and filter media replacement), resulting in unique removal mechanism patterns of RSF, GACF, and SSF (further discussed in “Microbial communities in biofilters and its potential functions in biological removal”) (Fig. 2).
In RSF, large amounts of nutrients are introduced into the filter bed, supporting rapid accumulation of the microbial community. This community includes nitrifying bacteria, methane-oxidising bacteria, iron-oxidising bacteria, and heterotrophic bacteria that were found to play important roles in degrading and removing ammonium, methane, and bentazone and in co-metabolising various organic micropollutants (such as CECs and naturally produced organic compounds) (de Vet et al. 2011; Wang et al. 2020). However, biomass accumulation (and also metal precipitates) in the filter leads to clogging and thus decreases the filtration rate over time (Maurya et al. 2020). To extend the life of RSF filters, backwashing (i.e. pumping water back into the filter media) has to be performed frequently every few hours up to days (flow rate varies from 1 to 20 m/h) (Hammes et al. 2011). As a result, biomass including microbial communities and their produced extracellular substances (EPS), and other residue compounds are promptly discarded from the filter, resulting in temporary loss of filter performance. Usually, RSF can be put back into operation instantly after cleaning (or within 15–60 min) (Brandt et al. 2017; Bruni and Spuhler 2012). This is due to biological removal only contributes to a limited extent in RSF (Fig. 2), and the effect of backwashing on RSF filter performance is usually not considered significant (Prevost et al. 2005). Owing to the introduction of higher nutrient concentrations from less treated influent water in RSF, recovery of the low biological removal contribution to its pre-cleaning state usually takes a shorter time.
In GACF and SSF, the filters are cleaned less frequently, i.e. GACF is backwashed weekly in summer and monthly in winter (Gibert et al. 2013; Knezev 2015); SSF is usually cleaned by scraping the SD layer every few months or even years (Schijven et al. 2013; Chan et al. 2018). Therefore, microbial communities can accumulate in the filters with less disturbance and contribute to biological removal. However, after applying the cleaning process, the recovery of the biological filter performance is, due to the relatively high ratio of biological removal contributions (Fig. 2), slower in GACF and SSF than RSF. Backwashing has been reported to have a significant impact on the community structure of biofilms accumulated on GACF media, causing a decrease in microbial diversity, and about half of the biomass can be washed away during backwashing (Liao et al. 2013a, 2015). Within a few days after backwashing and usually before the next backwash cycle, the biomass (probably related to biodegradation performance) returns to the pre-backwash concentration (Gibert et al. 2013; Liao et al. 2013a; Qi et al. 2019). In SSFs, the new Schmutzdecke layer takes some time to form and ripen after scraping. This usually takes a few days to weeks, and when the processed water can meet the requirement again, the filter can be used again for production (Burch and Thomas 1998; De Souza et al. 2021).
GAC has a high affinity for organic compounds because of the large surface area (500–1500 m2/g). GACF could physically filter pollutants through adsorption, attachment, and straining (Hijnen and Medema 2010; Knezev 2015). The flow rate controls the contact time between water and carbon, which is expressed as the Empty Bed Contact Time (EBCT). This parameter is widely used to estimate the removal performance of GACF. Previously, when the adsorption capacity of GAC is exhausted (i.e. expressed as a higher EBCT), the GAC is replaced or regenerated to maintain the desired removal performance of the GACF. With the recognition that increasing EBCT can improve biological removal, biofiltration features have been considered to contribute to the GACF performance (Brown et al. 2015; Oh et al. 2018). DWTPs that do not regularly change their filter media naturally change into biologically active GAC filters. At this stage, the adsorption capacity of the GAC is gradually depleted. At the same time, nutrients are continuously introduced into the filter to support the continued development of the microorganisms, which results in a rapid increase in the proportion of biological removal in the total removal contribution (Fig. 2) (Hammes et al. 2008; Velten et al. 2011; Brown et al. 2015). Regarding the BACF removal of organic carbon, chemical contaminants, and microbial contaminants (e.g. oocysts, bacteria, etc.), absorption removal is usually more efficient than biological removal due to the high-affinity properties of GAC materials. However, the biological removal specifically targets the biodegradable organic carbon fraction such as formaldehyde, geosmin, and compounds that are not identifiable, e.g. biodegradable humid substances (Hijnen and Medema 2010; Velten et al. 2011; Knezev 2015; Terry and Summers 2018). So far, limited predation of pathogens has been studied in GASF (Hijnen et al. 2007).
Differing from RSF and GACF, newly installed SSF filters cannot be used directly. SSF filters must be subjected to several months of ripening. During this ripening period, the so-called “Schmutzdecke”, which is a brown slimy layer or biofilm ranging from 0.5 to 2.0 cm (up to 3 cm) thick is formed on the surface of the filter (Matuzahroh et al. 2020; Ranjan and Prem, 2018). It is reported that sufficient biohazard removal efficiency (i.e. removal of pathogens from the water and reduction of nutrients causing poor biostability) can be achieved by a Schmutzdecke older than 6 months old (mature Schmutzdecke) to ensure that the treated water meets the desired quality requirements (Chaudhary et al. 2003; Bauer et al. 2011; Brandt et al. 2017).
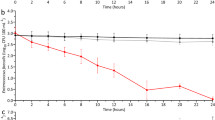
A mature Schmutzdecke layer consists of a diverse community of bacterial, archaeal, and eukaryotic microorganisms, EPS produced by the microorganisms, and other organic and inorganic debris and particulate matter (Pontius 2003; Wakelin et al. 2010). The mature Schmutzdecke layer is very biologically active, which increases water retention time, eliminates most of the contaminants including natural organic matter, transforms synthetic organic compounds, and retains pathogens from effluent water (Verma et al. 2017). Besides, trophic interactions in this layer also contribute to the bacterial removal (Haig et al. 2015b). Amongst all proposed and studied mechanisms, biological removal of contaminants has been found predominant in (mature) SSF, especially for removing pathogenic bacteria and viruses (Pfannes et al. 2015; Maurya et al. 2020). Removal of pathogenic bacteria and viruses cannot be effectively achieved by the physical straining of sand beds, as bacteria (0.1−10 µm) and viruses (0.01−0.1 µm) have small sizes compared to the pore size of SSF sand beds. A longer retention time of microbial contaminants in SSF filters (due to the slower flow rates) creates more opportunities for interactions with the indigenous microbial communities and their environment. For example, Haig et al. (2015b) firstly reported the evidence that E. coli removal in SSF is mainly caused by top-down trophic interactions, such as protozoan grazing and viral lysis.
Notably, the temperature and age of the Schmutzdecke were found to significantly affect the SSF removal efficiency, i.e. high temperature and matured age result in a better removal of microorganisms (Schijven et al. 2013; Haig et al. 2015a). This is consistent with the generally accepted assumption that the Schmutzdecke layer contributes most to the SSF removal efficiency, which highly associates with the activity of the complex microorganisms that inhabit the Schmutzdecke (Hijnen et al. 2004; Hijnen and Medema 2010; Pfannes et al. 2015; Schijven et al. 2013). However, viruses are considered a possible exception from the above assumption. Although some literature suggest that virus reduction could increase with Schmutzdecke maturation, most experimental data showed very limited or no virus removal in the Schmutzdecke layer (Bauer et al. 2011; Hijnen et al. 2004; Unger and Collins 2008). In the following sections, we will focus on the microbial community that has been revealed as providing the main responsibility for biological removal.
Microbial communities in biofilters and its potential functions in biological removal
It is wildly accepted that the composition, activity, and robustness of the biological community in biofilters determine the removal effectiveness of biofiltration units in DWTPs (Basu et al. 2015; Haig et al. 2015a; Oh et al. 2018). Previous studies about the ecology of drinking water biofilters are mainly focused on quantifying microbial communities, assessing microbial diversity, and discussing the potential function of those microorganisms present in biofilters (including potential pathogens).
Regarding the community composition, bacteria are predominantly present in all DWTP biofilters (including RSF, GAC, and SSF) (Wakelin et al. 2011; Bai et al. 2013; Oh et al. 2018) (Fig. 1b). Within the bacterial community, Proteobacteria, Planctomycetes, Acidobacteria, Bacteriodetes, and Nitrospirae are the core community (present at all DWTPs with a mean relative abundance >1%). The phylum Proteobacteria has the highest relative abundance in almost all drinking water biofilters, which might be linked to the capacity of certain Proteobacteria to thrive in systems with low dissolved organic carbon concentrations (Gerrity et al. 2018). These bacterial groups are also found in freshwater environments and can utilise various substrates (Newton et al. 2011). The bacterial community is considered the main carbon consumer amongst the whole microbial community (Oh et al. 2018). Overall, the bioactive nature of RSF, GACF, and SSF filters has been shown to relate to nitrifiers and iron-oxidising and manganese-oxidising bacteria (Cerrato et al. 2010; Feng et al. 2012; Brandt et al. 2017; Marcantonio et al. 2020).
Although bacteria are overall predominantly present in RSF, GACF, and SSF, biofilter community dissimilarity is still found between and within DWTPs (Lautenschlager et al. 2014; Gülay et al. 2016; Palomo et al. 2016; Oh et al. 2018; Poghosyan et al. 2020). In general, differences in community composition amongst DWTPs are greater than within DWTPs. Within DWTPs, microbial communities in the filter bed share more species with their effluents than influents (Ma et al. 2020; Guarin and Pagilla 2021). Different communities developed on different materials triggered a difference between effluent communities and affected the overall quality of the effluent water. In the following, the microbial community and the corresponding (potential) functions in RSF, GACF, and SSF are reviewed in detail.
Identity of microorganisms in biofilters and their potential functions
Rapid sand filtration
Bacteria were found to be the most abundant community members in RSF filters in both the filter material as well as the formed biofilms, while archaea were below 1%, and eukaryotic organisms only ranged from 4 to 7% (Table S2) (Bai et al. 2013; Gülay et al. 2016; Palomo et al. 2016).
At the phylum level, it was found that Proteobacteria and Nitrospirae were dominantly distributed in RSF filters followed by Acidobacteria and Bacteriodetes (Lautenschlager et al. 2014; Gülay et al. 2016; Palomo et al. 2016; Oh et al. 2018). The top three phyla Proteobacteria, Nitrospirae, and Acidobacteria build up most of the microbial community in RSF filters. Their abundance can be as high as 75−87 ± 18% of the total community (Gülay et al. 2016). The microbial community has been found with varying degrees of spatial heterogeneity in the filters (Palomo et al. 2016; Tatari et al. 2017). For instance, in a deep layer of RSF filters, Proteobacteria were more abundant (18.93 ± 1.31%) followed by Nitrospirae (9.22 ± 4.72%), while at the top of the same filters, Nitrospirae was found to be most abundant (26.08 ± 0.94%) followed by Proteobacteria (Palomo et al. 2016).
Within the microbial communities in RSF filters, the nitrifying guilds including ammonia-oxidising bacteria (AOB) (Nitrosomonas and Nitrosospira), ammonia-oxidising archaea (AOA) (Nitrosopumilus), and nitrite-oxidising bacteria (NOB) Nitrobacter and Nitrospira have been frequently observed and studied (Lautenschlager et al. 2014; Gülay et al. 2016; Palomo et al. 2016; Oh et al. 2018; Poghosyan et al. 2020). Tatari et al. (2017) reported a high abundance of Nitrospira (5−10% of the total community) consistently in RSF filters and up to 2 to 4 orders of magnitude more abundant than Nitrobacter and canonical AOBs, respectively. The high abundance of Nitrospira in nitrifying guilds is also reported by other studies (Poghosyan et al. 2020).
It was shown that the biological nitrification process is responsible for the reduction of the ammonia concentration in water in RSF filters (Oh et al. 2018; Poghosyan et al. 2020). The consistency of spatial dissimilarity of the ammonium removal capacity with the spatial heterogeneity of nitrifying guilds in RSF filters further supports the above hypothesis (Lee et al. 2014). Notably, the high abundance of Nitrospira spp., of which some species can oxidise ammonia completely to nitrate from many examined RSF filters reveals that non-canonical pathways for nitrification may dominate the RSF filters (Tatari et al. 2017; Oh et al. 2018; Vignola et al. 2018; Poghosyan et al. 2020), although their relative contribution to NH4 removal in DWTPs has not been examined yet.
Distribution of microorganisms associated with the biological oxidation of iron (de Vet et al. 2011; Li et al. 2013), manganese (Cerrato et al. 2010), arsenite (As(III)) (Gude et al. 2018), sulphur (Poghosyan et al. 2020), and methane (Terry and Summers 2018; Poghosyan et al. 2020) has also been observed in RSF filters, indicating the occurrence of their corresponding biological process and possible contribution to the RSF removal performance.
Granular activated carbon filtration
A high abundance of bacterial genera belonging to the Betaproteobacteria (61–80%), Alphaproteobacteria (25–43%), and Acidobacteria (7–14%) was found in GAC filter communities (Table S4) (Magic-Knezev et al. 2008; Knezev 2015; Oh et al. 2018). Members of the genera Bradyrhizobium (15%), Rhodopseudomonas (3.9%), and Afipia (2.5%), all belonging to the family Bradyrhizobiaceae, were dominantly present in GAC filters based on the metagenomic and 16S amplicon sequencing reported by Oh et al. (2018) and Lautenschlager et al. (2014). In addition, Polaromonas, Hydrogenophaga, Sphingomonas, Methylobacterium, and Variovorax were commonly isolated from the GAC filters, and their importance in biodegradation processes has been studied (Magic-Knezev et al. 2008; Wang et al. 2013; Zhou et al. 2021). Polaromonas organisms isolated from GACF were observed to be able to multiply at very low concentrations of carboxylic acids (Magic-Knezev et al. 2008). A Polaromonas strain JS666 isolated from a GACF has been observed associated with a potential utilisation of halogenated alkanes, cyclic alkanes, and (poly)aromatic compounds (Mattes et al. 2008). Members of the genus Hydrogenophaga can degrade methyl tert-butyl ether (MTBE), while members of the genus Sphingomonas are capable of degrading a wide range of xenobiotic compounds, including pesticides and micropollutants (i.e. terpene 2-methylisoborneol (MIB), isoproturon, polycyclic aromatic hydrocarbons (PAHs), and lindane) (Stolz et al. 2000; Kyselková et al. 2019; Abu Hasan et al. 2020). Variovorax organisms have been identified as bio-degraders of diverse aromatic compounds, including CECs ibuprofen (Murdoch and Hay 2015).
To further investigate the biodegradation functions of the microbial community in biofilters, Oh et al. (2018) reconstructed the metabolic pathways of biofiltration through metagenome analysis. The metabolic pathway associated with the degradation of aromatics was found to be significantly enriched in the GACF community compared to the other sand biofiltration processes. Members of the Rhizobiales (i.e. Bradyrhizobium, Afipia, Rhodopseudomonas, and Rhizobium) are the major groups encoding the aromatics degradation pathways amongst the total GACF community (Oh et al. 2018). It is likely that aromatics-bound DOCs are primarily biodegraded in GAC filters. The Rhizobiales (i.e. Bradyrhizobium) potentially play a key role in removing aromatic natural organic matter (NOMs) from the water during GACF (Oh et al. 2018).
Slow sand filtration
The microbial community of SSF filters is diverse both phylogenetically and metabolically (Haig et al. 2015a). Eukaryotes, archaea, and bacteria are all present in mature SSF filters (Table S6). Oh et al. (2018) investigated the microbial community in full-scale SSF filters with a combination of 16S rRNA gene sequencing and metagenomic analysis. The results showed that the majority of small subunit (SSU) rRNA gene sequences were phylogenetically affiliated to bacteria, while only < 1% and 2% of the SSU rRNA gene sequences were affiliated to eukaryotes and archaea, respectively.
The predominant bacterial groups have been extensively investigated (Wakelin et al. 2011; Oh et al. 2018), which are reviewed below. Members of the following bacterial phyla were abundant in various SSF filters: Proteobacteria (30−80%), Actinobacteria (1.2−16%), Acidobacteria (3–22%), Planctomycetes (4.4−14.9%), Nitrospirae (0−6%), and Bacteroidetes (4−25%) (Wakelin et al. 2011; Haig et al. 2014; Lautenschlager et al. 2014; D'Alessio et al. 2015; Li et al. 2017; Oh et al. 2018; De Souza et al. 2021). Proteobacteria including Alpha-, Beta-, and Gammaproteobacteria are always predominantly present in mature SSF filters (D’Alessio et al. 2015; Haig et al. 2014; Lautenschlager et al. 2014; Li et al. 2017; Oh et al. 2018; De Souza et al. 2021; Wakelin et al. 2011). This may be explained by the wide distribution of Proteobacteria bacteria in water sources and the variability of their metabolism (Newton et al. 2011; Li et al. 2017). Alphaproteobacteria are competitive at low nutrient concentrations (such as in river water) (Newton et al. 2011). The bacterial genus Bradyrhizobium belonging to the Alphaproteobacteria was found to be predominant in the SSF (Oh et al. 2018). They can contribute to nitrogen fixation and play a key role in removing aromatic NOMs in the SSF filter. Members of the Betaproteobacteria encompass a variety of methylotrophic and chemolithotrophic species, i.e. Methylobacillus and Methylophilus (D’Alessio et al. 2015). Phototrophic, chemotrophic, or chemolithotrophic bacteria belonging to the Gammaproteobacteria class (e.g. Methylococcales, Xanthomonadales, Chromatiales) were shown to be able to degrade complex organic compounds in SSF Schmutzdecke samples (Newton et al. 2011; Haig et al. 2015b).
The other predominant phyla present in SSF filters all belong to organic matter degradation-associated bacterial phyla (Liao et al. 2013b; Lautenschlager et al. 2014; D'Alessio et al. 2015; Haig et al. 2015a). For example, members of the Actinobacteria are commonly found in freshwater habitats where they play important roles in the degradation of organic compounds (Zhai et al. 2017). In addition, members of Streptomyces sp. can metabolise various compounds including sugars, amino acids, and aromatic compounds (Madigan et al. 2008). The genus Ferruginibacter belonging to the phylum Bacteroidetes has frequently been detected in water treatment plants, which are capable of hydrolysing organic matter (Zhai et al. 2017). In general, the bacterial community is the main consumer of soluble organic matter from the water (Oh et al. 2018). In addition to the above described predominant bacteria community, Gallionella, Leptothrix, and Crenothrix were observed and identified as the main microbial group responsible for iron and manganese oxidation in SSF (Demir 2016). The Nitrospira genus, belonging to nitrifying guild members, is found to be one of the dominant bacterial genera in various SSF (Oh et al. 2018; De Souza et al. 2021).
A low abundance of Archaea is observed in SSF filters (Wakelin et al. 2011; Oh et al. 2018). In a previous study, the archaeal community of the Schmutzdecke was dominated by aerobic chemo-heterotrophically Euryarchaeota, consisting mostly of Halobacteriales (photo-autotrophic taxa was excluded due to the dark environment of SSF) (Wakelin et al. 2011). Wakelin et al. (2011) hypothesised that archaea are active in the removal of dissolved organic carbon from the influent water.
Regarding the eukaryotic community, protists, protozoa, green alga, and fungi are found in SSF filters (Wakelin et al. 2011). The authors noted that the protists and protozoa are closely involved in the removal of microbial contaminants (e.g. pathogenic bacteria). This result is in line with the results found by Oh et al. (2018), who reported that the majority of the eukaryotic sequences from SSF filters were taxonomically affiliated with Animalia (54%) and Viridiplantae (20%). In particular, earthworm populations Enchytraeidae belonging to Animalia were enriched in the surface of the Schmutzdecke layer of the SSF filter. These earthworms have been found in other filter biofilms, and they can control the bacterial population by grazing (Lourenço and Nunes 2017). Notably, even at low abundance, eukaryotic communities in the SSF are strongly related to the removal of biological contaminants (e.g. through predation and grazing) (Weber-Shirk and Dick 1997; Stott et al. 2001; Haig et al. 2015b). However, a comprehensive analysis of the eukaryotic community in the Schmutzdecke of SSF is still limited.
Regarding temporal changes, fluctuations are mostly observed when a SSF is newly installed. The abundance, biodiversity, and also evenness of the microbial community in SSF filters increase with the increasing filter age (Ramond et al. 2013; Haig et al. 2015a). When the filter becomes mature, the temporal changes are marginal.
As for the spatial distribution, researchers proposed that vertical (depth) variation in the sand filter should be present in the SSF, as it relates to the chemical gradients that drive changes in community composition (Lin et al. 2012; Haig et al. 2014). However, this presumption has been challenged recently after the extensive utilisation of advanced microbial community analysis approach, i.e. 16S rRNA amplicon sequencing and metagenomics sequencing (Haig et al. 2015a; Wakelin et al. 2011; Li et al. 2017). Wakelin et al. (2011) noted that, although the highest amount of DNA content was found in the surface of the Schmutzdecke and declined along with depth, the overall community composition structure was similar through the filter depth for bacteria, archaea, and eukaryotes. Another study showed that bacterial communities of sand samples from different depths were similar in two full-scale SSFs (Haig et al. 2015a). Li et al. (2017) reported that the bacterial communities are uniform from the surface to the middle part of SSF filters with high stability. Nevertheless, further study is required to determine whether chemical gradients in water exist which might affect the SSF microbial communities (Haig et al. 2015a).
Abiotic and biotic factors affecting the assemblage of microbial communities in biofilters
Abiotic factors
A significant effect of media material type on bacterial community composition in the biofilter has been observed (Gerrity et al. 2018; Oh et al. 2018; Vignola et al. 2018; Ma et al. 2020). This is due to the different properties of various filter media, such as intraparticle porosity, surface area, chemical properties, and adsorption capacity. Gerrity et al. (2018) and Vignola et al. (2018) concluded that GACF biofilter communities tend to be more phylogenetically diverse than those on other filter media types like anthracite and sand. Vignola et al. (2018) studied the development of microbial communities on sand filters and GAC, using the same source water in laboratory-scale columns. The results show that Planctomyces (11−13%) and Gemmata (1.3−8.1%) were the most abundant genera in the sand filters. In contrast, in GAC filters, the two highest relative abundance of operational taxonomic units (OTUs) on GAC filters were unidentified beyond the Gammaproteobacteria class level (5.6−6.2%). Species belonging to the genus of Gordonia, Sulfuritalea, and Nitrospira were identified as main contributors in biofiltration. They are highly abundant in sand filters but have a very low abundance in GAC filters. Significant community differences were also observed between the GAC and anthracite filters (Ma et al. 2020). In addition to filters from independent DWTPs (including studied lab-scale and pilot-scale plants), the above patterns are also consistent in different filters from the same DWTP. Lautenschlager et al. (2014) and Oh et al. (2018) studied the microbial community of biofilters including RSF, GACF, and SSF within a full-scale DWTP. The results indicated that Bradyrhizobiaceae were more abundant in GAC filters, whereas Nitrospira were enriched in the sand-associated filters (RSF and SSF).
In addition to the filter media, Oh et al. (2018) suggested that the influent water is another important factor contributing to the differences in bacterial communities of biofilters from the same DWTP. Organic carbon is removed disproportionately at each filtration stage and the GAC receives ozonated water; therefore, substances in the RSF and GACF are more easily degraded than in the SSF filter; as a result, different influent water affects the native biological community of the filter (Oh et al. 2018).
Nutrient levels such as dissolved organic carbon (DOC), ammonia nitrogen, and phosphorus in the influents could affect the bacterial diversity and community composition in biofilters (Li et al. 2010; Liao et al. 2013a; Gerrity et al. 2018; Knezev 2015). Ma et al. (2020) reported that although the influence is tempered by filter design and operating conditions (e.g. filter type, backwash) (De Souza et al. 2021), microbiomes in biofilters are primarily shaped by water quality conditions. This matches the finding of a high similarity in microbial taxa on all treatment plant filters from the same DWTPs (Lautenschlager et al. 2014). Filters operating with ozonated water generally contain significantly higher biomass levels than in the same system operating with non-ozone water. The number of different genera within the Betaproteobacteria was higher in GAC filters treated with ozonated water than in filters treated with non-ozone water (Knezev 2015). In addition, Rhizobiales were more frequently detected in GAC filters that received ozonated water. Li et al. (2010) found that the addition of phosphorus significantly changed the genus compositions of Betaproteobacteria in BAC reactors. The authors also found that the richness and evenness of the overall microbial community were decreased in all bioreactors when adding phosphorus. Trace amounts of organic compounds in influent water, including personal care products, household chemicals, and pharmaceutically active compounds (PhACs), have been reported to significantly alters the microbial community in biofilters. Within the Schmutzdecke, the relative abundance of Proteobacteria increased approximately from 30 to 99%, while the occurrence of Bacteroidetes dropped from 37 to 1% during the study (D’Alessio et al. 2015). These PhAC-induced microbial community changes interfered with the bacterial removal performance of SSF in Schmutzdecke, decreasing from 95% to less than 20%. Furthermore, Delgado-Gardea et al. (2019) demonstrated that additional brass in influent water changed the microbial community structure of the Schmutzdecke from a bacteria-dominated community to a eukaryote-dominated community (Delgado-Gardea et al. 2019). The brass-SSF filter had the eukaryotic Streptophyta dominating (31.4%), followed by Gluconobacter (24.6%) and Acetobacter family (7.7%); these genera were absent in all other SSF treatments from the same study.
RSF and GACF communities potentially select fast-growers differing from SSF communities, which is consistent with the observed highest rate of dissolved organic matter removal by RSF and GACF (Lautenschlager et al. 2014; Oh et al. 2018). This may associate with the effects of flow rates or EBCT. In other words, compared to filters operated under slow flow rates, a higher flow rate results in relatively short bed contacting time (lower EBCT) and therefore selects fast-growers in rapid filters.
In mature biofilters, the temperature had a non-significant impact on total biomass levels. However, the biological activity of the biotic community at high temperatures could be significantly higher than at low temperatures. This phenomenon is observed from season fluctuation in various biofilters. For instance, through tetrazolium reduction assays, Fonseca et al. (2001) reported that specific dehydrogenase activity was 70% higher in systems operated at ambient temperatures (> 12 °C) than in the systems held at 3 °C.
In addition to operational conditions, the biofilter scales can also affect the community composition. Drastically different responses in microbial community structure were detected in bench-scale and pilot-scale BAC reactors after phosphorus addition (Li et al. 2010). In that study, the relative abundance of perchlorate-reducing bacteria (PRB) Dechloromonas (the only known PRB in the system) was observed to increase from 15.2 to 54.2% in the bench-scale but decreased from 7.1 to 0.6% in the pilot-scale reactor, while Zoogloea increased from 17.9 to 52.0%.
Biotic factors
The microbial community in influent water may affect the community in filters. This effect has been frequently reported. Certain species that inhabit freshwater or influent water have a better capacity to attach and survive in biofilms of the biofilters. However, it is found that filter communities are strongly assembled by selection rather than immigration (Vignola et al. 2018). This finding is consistent with the limited effect of “bioaugmentation” in drinking water biofilters.
Bioaugmentation is an eco-friendly and economically viable method to enhance the degradation of pollutants and pathogens by adding pre-grown functional microorganisms or microbial symbionts to the media or environment. It was initially applied in wastewater treatment plants (WWTPs) and now is receiving increasing attention in DWTPs (Herrero and Stuckey 2015).
Albers et al. (2018) successfully primed nitrification in RSF filters via bioaugmentation of nitrifying communities from an existing biofilter enriched on quartz sand. Adding nitrifying community inoculum substantially decreased the lag time before nitrification commenced in the new RSF filter. However, the bioaugmented microorganisms were eventually outcompeted by native nitrifiers (Albers et al. 2018). This phenomenon is similar to the long-term loss of bioaugmented microorganisms observed in other drinking water studies (Davidson et al. 2011). It is consistent with other bioaugmented studies where a very limited effect of inoculant on the assemblage of microbial community has been found, although adding inoculant could shorten the ripening time in some biofilters (Davidson et al. 2011; Bai et al. 2016; Chan et al. 2018; Breda et al. 2019).
Several factors are hypothesised to contribute to the loss of inoculated cells from the sand filters, such as starvation due to too low levels of nutrient and assimilative organic carbon (AOC), mass-transfer limitations, antagonism by indigenous microorganisms in the filters, or simply continuous washout from the filter (Horemans et al. 2017; Albers et al. 2018). In general, the family-level composition was convergent even across different inoculants observed in other multi-replicated enrichment communities studies (Estrela et al. 2021).
Correlation of microbial community and filtration performance
In this section, all three filtration processes are included; however, the focus will be on SSF as biofiltration plays a major role there. Although the successful function of biofiltration relies on the interaction amongst various microbial communities, especially in SSF (Wakelin et al. 2011), the correlation between the community composition and filtration efficiency (for removing chemical and biological hazards) remains unclear. Studies show that a similar filtration performance could be obtained from filters with significantly different community compositions and structures (Li et al. 2010; Vignola et al. 2018). For example, Li et al. (2010) studied the biological activated carbon (BAC) filtration in bench-scale and pilot-scale reactors operating under similar phosphorus addition conditions. After adding phosphorus, significantly different responses from the microbial community composition were observed. However, these bench and pilot scales corresponded to a similar filtering performance regarding perchlorate and nitrate removal (Li et al. 2010). In addition, Delgado-Gardea et al. (2019) reported that the bacterial removal efficiency was not significantly affected by metals, even though the brass changed the microbial community structure of the Schmutzdecke from a bacteria-dominated community to a eukaryote-dominated community (Delgado-Gardea et al. 2019).
To address this contradicting observation, researchers are trying to find out the core microbiome that is involved in biological removal processes. Some bacterial genera associated with SSF filter performance have been reported by Haig et al. (2015a). In this study, the filter performance was monitored by testing 15 water quality parameters (including ammonia, coliforms, DOC, nitrate, nitrite, total number of viable bacteria). Afterwards, the performance was further evaluated by an aggregate performance metric (Haig et al. 2014). It was found that the abundance of Sphingomonas and Halomonas (belonging to the Proteobacteria), and Acinetobacter (belonging to the Actinobacteria) increased during periods of better removal performance. In contrast, when the SSF filter showed poor performance, the abundance of Acidovorax and Sphingobium having similar biodegradation functions as Sphingomonas and Acinetobacter was noticed, which is explained by niche competition (Haig et al. 2015a).
From the same study, a strong positive correlation was found between species evenness and filter performance, which is in line with “the insurance hypothesis” proposed by Yachi and Loreau (1999). The hypothesis stated: “biodiversity ensures ecosystems against declines in their functioning because many species provide greater guarantees that some will maintain functioning even if others fail”. Considering that the microbial community of bioactive filters could affect and even shape the microbial community of the effluent water (Haig et al. 2014; Lautenschlager et al. 2014; Li et al. 2017; Chan et al. 2018), it seems that biostability in drinking water also benefits from a large diversity of less active species, rather than of a few very active ones. This phenomenon may only be observed in slow sand filters (i.e. SSF), while in rapid sand filters (i.e. RSF) and GACF, some stable dominant species can be more relevant because of the short reaction time.
In addition to the positive effect of communities in filters, the release of members of the microbial community from sand filters into the effluent water has been noticed (Bichai et al. 2010, 2014). If persistent pathogenic microorganisms survive in the filter or survive ingestion by protozoa, they can be released from the biofilter into the drinking water. Opportunistic pathogens were identified by 16S rRNA-based analysis, and the results suggest that the Schmutzdecke indeed contains opportunistic pathogens causing human infections (Lautenschlager et al. 2014). These findings pose a risk to drinking water safety, especially for DWTPs that use SSFs as the final treatment step. Therefore, time-course monitoring of opportunistic pathogens in the Schmutzdecke is necessary to ensure consistent drinking water quality.
Status and challenges of the study of biofiltration in drinking water production
Approaches using descriptive and inductive measurement with correlation analysis have been widely performed in drinking water biofilter studies (Kirisits et al. 2019). As an example, the microbial community of biofilters is investigated by the sequencing of the 16S rRNA or functional genes. After grouping sequences into arbitrary OTUs, amplicon sequence variant (ASV), or bins (specific for metagenomic sequencing), statistical methods are used to identify the diversity within and amongst samples or sample groups. Afterward, quantitative correlation analysis is conducted for phylotype relative abundance and biofilter characteristics (e.g. a specific hazard group removing performance), focused on correlation expected to be statistically significant. Subsequently, induction and infer methodology are invoked to link the observed community from biofilters to the known (potential) functional communities. The resulting outcomes will be compared to literature data to find support for inference. This example is a typical top-down study approach (Faust 2019), and it remains at the descriptive level because of delivering associations rather than causal relationships.
Regarding the descriptive resolution, almost all molecular techniques including 16S rRNA and metagenome sequencing for studying microbial communities can only identify the species level but not the strain level (Prosser 2020). This limitation can lead to misleading conclusions, as the function and phenotypic characteristics can be dramatically different amongst strains from the same species. To link bio-community to specific ecological functions, bioinformatics tools such as Tax4fun (Aßhauer et al. 2015; Wemheuer et al. 2020) and PICRUST (Langille et al. 2013; Douglas et al. 2020) pipelines have been developed. The resulting 16S rRNA gene data with gene data repositories can be used to predict metagenomes (Koo et al. 2017; Li et al. 2017). For example, when ammonia oxidation is studied, the presence and amount of functional gene amoA encoding a subunit-A of the ammonia monooxygenase are investigated (Lee et al. 2014). However, the presence of the corresponding functional genes does not mean that these genes are expressed to contribute to the specific biodegradation/biofiltration process. In addition, the databases used by these bioinformatics tools (e.g. NCBI RefSeq database, Greengenes database) are mostly populated with medical/human-related sequences (McDonald et al. 2012; O’Leary et al. 2016; Koo et al. 2017). Therefore, for samples outside the biomedical domain, such as samples from DWTPs scenarios, inference from the default database may be not accurate.
Furthermore, to date, almost all studies on microbial communities of drinking water filters have focused on the total community via DNA-based sequencing methods. The total community (DNA) cannot distinguish the active microorganisms from dormant or dead community members, which could lead to a biased outcome. For example, significant differences between the total and active community in the drinking water distribution system (DWDS) have been noticed by Inkinen et al. (2016). In this study, the active (RNA) bacterial community consists of Gammaproteobacteria (Pseudomonas spp., Yersinia spp.) and Abditibacteriota (FBP) phyla in the biofilm from a circulating hot water system. This active community largely differed from the total community composed of mainly Betaproteobacteria (Limnohabitans sp., Methylotenera sp., Comamonadaceae family) and Actinobacteria (Corynebacterium spp.). Dormant or dead microorganisms do not contribute to the biodegradation/biofiltration process and so can give a biased view on the biofiltration processes. To truly understand and predict environmental processes, the distinction between active, inactive, and dead microbial cells is crucial (Singer et al. 2017). To address this issue, RNA-based techniques (such as reverse transcription (RT)-PCR) are suggested to complement DNA-based sequencing. The study of Inkinen et al. (2019) gave a more complete picture of the microbial community of biofilters by incorporating both DNA- and RNA-based sequencing methods. Furthermore, combining Stable Isotope Probing (SIP) with 16S Amplicon and metagenomics sequencing could help to further the understanding of the removal processes and the in situ trophic interactions (Haig et al. 2015b; Singer et al. 2017; Kleiner et al. 2018).
On the other hand, the bacterial group is always the main focus owing to its high abundance in the community, while the eukaryotic groups are overlooked from some studies (Lautenschlager et al. 2014; Gude et al. 2018). The importance and contribution of a microbial group should not be deduced from abundance alone, since low-abundance keystone species can play major roles in ecosystem function. A recent study of sand biofilters during manganese load fluctuations reveals that the keystone species in the ecosystem could be rare (Zhao et al. 2021). In that study, keystone microorganisms contributing to the ecological stability and Mn(II) oxidation in sand biofilters were identified by combing microbiome network and functional modules analyses. The relatively rare species Candidatus Entotheonella palauensis (relative abundance 0.39%) was identified as a module hub and it was presumed as one of the keystone species. In contrast, although the well-known manganese-oxidising bacterium Hyphomicrobium dominated the sand biofilter at all depths (relative abundance 3.3–12.7%), it exhibited only a few microbial interactions in the symbiotic network. This finding indicates that implying taxa with low abundance can play important roles in ecosystem function and should not be neglected. Regarding eukaryotic communities, even at low abundance, they may be closely correlated to the removal of biological contaminants through biological activities such as grazing or predation in the Schmutzdecke (Weber-Shirk and Dick 1997; Stott et al. 2001; Haig et al. 2015b). Therefore, a comprehensive analysis of the eukaryotic groups should be performed in future studies.
Outlook on the microbial ecology of biofilters
To obtain a comprehensive understanding of the microbial ecology of biofiltration and promote its application to meet today's growing challenges of safe drinking water production, future studies are recommended to answer the following key questions: (i) who are the active “keystone species” in the biofilters? (ii) what bioprocess actively occurs in-situ during biofiltration? (iii) how does the bioprocess contribute to the filtration performance? and (iv) how to modify the assemblage of the indigenous community (through operational parameter optimisation and/or bioaugmentation, etc.) in biofilters to achieve the target biofiltration efficiency?
To address these important questions, we suggest studying both DNA and RNA in microbial community analysis. In addition, apart from the study of bacteria, eukaryotes should also be studied. We also recommend to further investigate the link of microbial diversity to specific ecological functions and reveal functional genes from the microbial community involved in metabolic and catabolic pathways. Several techniques and approaches such as meta-omics (e.g. transcriptomics analysis), nanoscale secondary ion mass spectrometry (NanoSIMS), and confocal laser scanning microscopy-fluorescent in situ hybridisation (CLSM-FISH) can be used for studying the microbial ecology of biofilters (Gebert et al. 2008; Jehmlich et al. 2016; Lueders et al. 2016; Musat et al. 2016; Singer et al. 2017; Barlow et al. 2020; Berg et al. 2020). In addition, proteomics studies and specific enzyme activity analysis are proposed to assist the functional study (Douterelo et al. 2014; Kleiner et al. 2017, 2018; Oh et al. 2018; Kumar et al. 2019; Barlow et al. 2020). Regarding the enzyme activity, the choice of the specific enzyme depends on the research question and previous findings. For example, Lautenschlager et al. (2014) observed that polysaccharides can be better degraded in the SSFs than in GAC filters and RSFs, and the authors suspected this was due to the activity of extracellular enzymes. Regarding the wish to have fewer clogging problems during biofiltration, it would be valuable to study the polysaccharide-degrading enzymes in biofilters. In addition, nitrate reductase (Nar) and nitrite reductase (Nir) were stated as the key enzymes in a biofilter that were associated with a better denitrification performance (Jia et al. 2022). Cao et al. (2022) reported that enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase was strongly correlated with individual trace organic chemical transformation in the water biofiltration process.
Reviewing the current biofiltration developments in drinking water treatment systems, we observed that most studies were carried out individually, and only a few snapshots of microbial community data are available at various taxonomy levels in the specific DWTPs. To make sequencing outcomes from various studies comparable, and also understand their functional meaning in the specific ecosystem more straightforward, work similar to that of the activated sludge “MiDAS (Microbial Database of Activated Sludge)” project is proposed. Within the MiDAS project, an ecosystem-specific platform for wastewater treatment systems has been established, in which standardised wet-lab protocols and a comprehensive activated sludge microbial database of complete 16S rRNA gene sequences (MiDAS) have been discussed and developed (Dueholm et al. 2022). We believe this type of work is also valuable in drinking water ecosystems, providing a platform for generating a more comprehensive understanding of microbial ecology in drinking water production.
Furthermore, more attention should be paid on the validation and application of biofiltration research outcomes in the production of actual drinking water from a broader view. The final application aim of the biofiltration study is to develop and maintain “Good-performance” biofilters that (1) have a higher removal efficiency for target contaminants (various depending on the biofilter type, see Fig. 1c); (2) are robust to environmental fluctuation; and (3) are resistant to clogging and pathogens release problems. However, most biofiltration studies only consider one of the perspectives mentioned above. Considering the contribution of the microbial community in the biofiltration process, a comprehensive multi-dimensional model to describe the microbe community effects on all “good performance” perspectives is needed. With this model being established, a concise definition of the biofilter “healthy microbiome” may be able to be developed and be used to monitor the health status of running biofilters. For example, if the “high evenness” is included in the “healthy microbiome” definition and “keystone species” can be identified, timely monitoring of the evenness of the biofilter microbiome and detecting the abundance of the “keystone species” group via q-PCR could be included into the routine monitoring system. When the filter microbial community shows “high evenness”, and the “keystone species” can be regularly detected, the biofilter can be diagnosed as a “healthy filter”. In contrast, when the evenness and the abundance of “keystone species” are below a certain standard, an alarm should be voiced out. And the risk groups of microorganisms that may result in an unhealthy biofilter, so-called disease risky microbiome, should be further tested. In these ways, an economic and rapid solution for monitoring the “health” status of biofilters and preventing accidental outbreaks can be developed. In addition, the “pro- or prebiotics treatment” idea, regulating the filter microbial community to the status by feeding certain live microbial groups and/or nutrients, holds great promise for modulating the health status of biofilters and drinking water risk management.
To summarise, the black box of the biofiltration process in drinking water has not been illuminated yet. Future studies about “who is there?” should be conducted in the way of eliminating group biases (i.e. not only the bacteria group), and new studies answering questions such as “who is doing what?” should be focused on revealing the cause-relations between the microbial community and removal performance. Furthermore, to better apply the biofiltration research outcomes in real production, a comprehensive model describing this correlation is urgently needed. It is sincerely hoped that outcomes of biofilters studies could extensively contribute to developing safer, more reliable, and more sustainable DWTPs.
References
Abu Hasan H, Muhammad MH, Ismail NI (2020) A review of biological drinking water treatment technologies for contaminants removal from polluted water resources. J Water Process Eng 33:101035. https://doi.org/10.1016/j.jwpe.2019.101035
Albers CN, Ellegaard-Jensen L, Hansen LH, Sørensen SR (2018) Bioaugmentation of rapid sand filters by microbiome priming with a nitrifying consortium will optimize production of drinking water from groundwater. Water Res 129:1–10. https://doi.org/10.1016/j.watres.2017.11.009
Aßhauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31:2882–2884. https://doi.org/10.1093/bioinformatics/btv287
Asami T, Katayama H, Torrey JR, Visvanathan C, Furumai H (2016) Evaluation of virus removal efficiency of coagulation-sedimentation and rapid sand filtration processes in a drinking water treatment plant in Bangkok, Thailand. Water Res 101:84–94. https://doi.org/10.1016/j.watres.2016.05.012
Bai Y, Chang Y, Liang J, Chen C, Qu J (2016) Treatment of groundwater containing Mn(II), Fe(II), As(III) and Sb(III) by bioaugmented quartz-sand filters. Water Res 106:126–134. https://doi.org/10.1016/j.watres.2016.09.040
Bai Y, Liu R, Liang J, Qu J (2013) Integrated metagenomic and physiochemical analyses to evaluate the potential role of microbes in the sand filter of a drinking water treatment system. PLoS ONE 8:e61011. https://doi.org/10.1371/journal.pone.0061011
Barlow JT, Bogatyrev SR, Ismagilov RF (2020) A quantitative sequencing framework for absolute abundance measurements of mucosal and lumenal microbial communities. Nat Commun 11:1–13. https://doi.org/10.1038/s41467-020-16224-6
Basu OD, Dhawan S, Black K (2015) Applications of biofiltration in drinking water treatment - a review. J Chem Technol Biotechnol 91:585–595. https://doi.org/10.1002/jctb.4860
Bauer R, Dizer H, Graeber I, Rosenwinkel KH, López-Pila JM (2011) Removal of bacterial fecal indicators, coliphages and enteric adenoviruses from waters with high fecal pollution by slow sand filtration. Water Res 45:439–452. https://doi.org/10.1016/j.watres.2010.08.047
Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, Kazou M, Kinkel L, Lange L, Lima N, Loy A, Macklin JA, Maguin E, Mauchline T, McClure R, Mitter B, Ryan M, Sarand I, Smidt H, Schelkle B, Roume H, Kiran GS, Selvin J, de Souza RSC, van Overbeek L, Singh BK, Wagner M (2020) Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. https://doi.org/10.1186/s40168-020-00875-0
Bichai F, Barbeau B, Dullemont Y, Hijnen W (2010) Role of predation by zooplankton in transport and fate of protozoan (oo)cysts in granular activated carbon filtration. Water Res 44:1072–1081. https://doi.org/10.1016/j.watres.2009.09.001
Bichai F, Dullemont Y, Hijnen W, Barbeau B (2014) Predation and transport of persistent pathogens in GAC and slow sand filters: a threat to drinking water safety? Water Res 64:296–308. https://doi.org/10.1016/j.watres.2014.07.005
Bishaw D, Kebede F (1999) Evaluation on the efficiency of rapid sand filtration. Integrated development for water supply and sanitation: proceedings of the 25th WEDC Conference, Ethiopia
Brandt MJ, Johnson KM, Elphinston AJ, Ratanyaka DD (2017) Water Filtration (Chapter 9). In: supply, 7th edn. Elsevier, pp 367–406. https://doi.org/10.1016/b978-0-08-100025-0.00009-0
Breda IL, Søborg DA, Ramsay L, Roslev P (2019) Manganese removal processes during start-up of inoculated and non-inoculated drinking water biofilters. Water Qual Res J Canada 54:47–56. https://doi.org/10.2166/wqrj.2018.016
Brown J, Summers RS, Le Chevallier M, Collins H, Roberson JA, Hubbs S, Dickenson E (2015) Biological drinking water treatment? Naturally. J Am Water Works Assoc 107:20–30. https://doi.org/10.5942/jawwa.2015.107.0183
Bruni M, Spuhler D (2012) Rapid sand filtration. Sustainable Sanitation and Water Management Toolbox - SSWM.info. https://sswm.info/sswm-university-course/module-6-disaster-situations-planning-and-preparedness/further-resources-0/rapid-sand-filtration. Accessed 26 March 2022
Burch JD, Thomas KE (1998) Water disinfection for developing countries and potential for solar thermal pasteurization. Sol Energy 64(1–3):87–97. https://doi.org/10.1016/S0038-092X(98)00036-X
Cao L, Wolff D, Liguori R, Wurzbacher C, Wick A (2022) Microbial biomass, composition, and functions are responsible for the differential removal of trace organic chemicals in biofiltration systems: a batch study. Front Water 4:832297. https://doi.org/10.3389/frwa.2022.832297
Cerrato JM, Falkinham JO, Dietrich AM, Knocke WR, McKinney CW, Pruden A (2010) Manganese-oxidizing and -reducing microorganisms isolated from biofilms in chlorinated drinking water systems. Water Res 44:3935–3945. https://doi.org/10.1016/j.watres.2010.04.037
Chan S, Pullerits K, Riechelmann J, Persson KM, Rådström P, Paul CJ (2018) Monitoring biofilm function in new and matured full-scale slow sand filters using flow cytometric histogram image comparison (CHIC). Water Res 138:27–36. https://doi.org/10.1016/j.watres.2018.03.032
Chaudhary DS, Vigneswaran S, Ngo H-HH, Shim WG, Moon H (2003) Biofilter in water and wastewater treatment. Korean J Chem Eng 20:1054. https://doi.org/10.1007/BF02706936
D’Alessio M, Yoneyama B, Kirs M, Kisand V, Ray C (2015) Pharmaceutically active compounds: their removal during slow sand filtration and their impact on slow sand filtration bacterial removal. Sci Total Environ 524–525:124–135. https://doi.org/10.1016/j.scitotenv.2015.04.014
Davidson AN, Chee-Sanford J, Lai HY, Ho CH, Klenzendorf JB, Kirisits MJ (2011) Characterization of bromate-reducing bacterial isolates and their potential for drinking water treatment. Water Res 45:6051–6062. https://doi.org/10.1016/j.watres.2011.09.001
Delgado-Gardea MCE, Tamez-Guerra P, Gomez-Flores R, Garfio-Aguirre M, Rocha-Gutiérrez BA, Romo-Sáenz CI, Zavala-Díaz de la Serna FJ, Eroza-de la Vega G, Sánchez-Ramírez B, González-Horta M del C, Infante-Ramírez M del R (2019) Streptophyta and acetic acid bacteria succession promoted by brass in slow sand filter system Shmutzdeckes. Sci Rep 9:7021. https://doi.org/10.1038/s41598-019-43489-9
Demir NM (2016) Experimental study of factors that affect iron and manganese removal in slow sand filters and identification of responsible microbial species. Pol J Environ Stud 25:1453–1465. https://doi.org/10.15244/pjoes/62679
De Souza FH, Roecker PB, Silveira DD, Sens ML, Campos LC (2021) Influence of slow sand filter cleaning process type on filter media biomass : backwashing versus scraping. Water Res 189:116581. https://doi.org/10.1016/j.watres.2020.116581
de Vera GA, Lauderdale C, Alito CL, Hooper J, Wert EC (2019) Using upstream oxidants to minimize surface biofouling and improve hydraulic performance in GAC biofilters. Water Res 148:526–534. https://doi.org/10.1016/j.watres.2018.10.085
de Vet WWJM, Dinkla IJT, Rietveld LC, van Loosdrecht MCM (2011) Biological iron oxidation by Gallionella spp. in drinking water production under fully aerated conditions. Water Res 45:5389–5398. https://doi.org/10.1016/j.watres.2011.07.028
Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MG (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. https://doi.org/10.1038/s41587-020-0548-6
Douterelo I, Boxall JB, Deines P, Sekar R, Fish KE, Biggs CA (2014) Methodological approaches for studying the microbial ecology of drinking water distribution systems. Water Res 65:134–156. https://doi.org/10.1016/j.watres.2014.07.008
Dueholm MKD, Nierychlo M, Andersen KS, Rudkjøbing V, Knutsson S, Albertsen M, Nielsen PH (2022) MiDAS 4: a global catalogue of full-length 16S rRNA gene sequences and taxonomy for studies of bacterial communities in wastewater treatment plants. Nat Commun 13:1908. https://doi.org/10.1038/s41467-022-29438-7
Estrela S, Sánchez Á, Rebolleda-Gómez M (2021) Multi-replicated enrichment communities as a model system in microbial ecology. Front Microbiol 12:657467. https://doi.org/10.3389/fmicb.2021.657467
Faust K (2019) Towards a better Understanding of microbial community dynamics through high-throughput cultivation and data integration. mSystems 4:e00101-19. https://doi.org/10.1128/msystems.00101-19
Favere J, Barbosa RG, Sleutels T, Verstraete W, De Gusseme B, Boon N (2021) Safeguarding the microbial water quality from source to tap. NPJ Clean Water 4:28. https://doi.org/10.1038/s41545-021-00118-1
Feng S, Xie S, Zhang X, Yang Z, Ding W, Liao X, Liu Y, Chen C (2012) Ammonium removal pathways and microbial community in GAC-sand dual media filter in drinking water treatment. J Environ Sci (china) 24:1587–1593. https://doi.org/10.1016/S1001-0742(11)60965-0
Fonseca AC, Scott Summers R, Hernandez MT (2001) Comparative measurements of microbial activity in drinking water biofilters. Water Res 35:3817–3824. https://doi.org/10.1016/S0043-1354(01)00104-X
Gebert J, Stralis-Pavese N, Alawi M, Bodrossy L (2008) Analysis of methanotrophic communities in landfill biofilters using diagnostic microarray. Environ Microbiol 10:1175–1188. https://doi.org/10.1111/j.1462-2920.2007.01534.x
Gerrity D, Arnold M, Dickenson E, Moser D, Sackett JD, Wert EC (2018) Microbial community characterization of ozone-biofiltration systems in drinking water and potable reuse applications. Water Res 135:207–219. https://doi.org/10.1016/j.watres.2018.02.023
Gibert O, Lefèvre B, Fernández M, Bernat X, Paraira M, Calderer M, Martínez-Lladó X (2013) Characterising biofilm development on granular activated carbon used for drinking water production. Water Res 47:1101–1110. https://doi.org/10.1016/j.watres.2012.11.026
Greenstein KE, Lew J, Dickenson ERV, Wert EC (2018) Investigation of biotransformation, sorption, and desorption of multiple chemical contaminants in pilot-scale drinking water biofilters. Chemosphere 200:248–256. https://doi.org/10.1016/j.chemosphere.2018.02.107
Guarin TC, Pagilla KR (2021) Microbial community in biofilters for water reuse applications: a critical review. Sci Total Environ 773:145655. https://doi.org/10.1016/j.scitotenv.2021.145655
Gude JCJ, Rietveld LC, van Halem D (2018) Biological As(III) oxidation in rapid sand filters. J Water Process Eng 21:107–115. https://doi.org/10.1016/j.jwpe.2017.12.003
Gülay A, Musovic S, Albrechtsen HJ, Al-Soud WA, Sørensen SJ, Smets BF (2016) Ecological patterns, diversity and core taxa of microbial communities in groundwater-fed rapid gravity filters. ISME J 10:2209–2222. https://doi.org/10.1038/ismej.2016.16
Haig SJ, Collins G, Davies RL, Dorea CC, Quince C (2011) Biological aspects of slow sand filtration: past, present and future. Water Sci Technol Water Supply 11:468–472. https://doi.org/10.2166/ws.2011.076
Haig SJ, Quince C, Davies RL, Dorea CC, Collins G (2014) Replicating the microbial community and water quality performance of full-scale slow sand filters in laboratory-scale filters. Water Res 61:141–151. https://doi.org/10.1016/j.watres.2014.05.008
Haig SJ, Quince C, Davies RL, Dorea CC, Collinsa G (2015a) The relationship between microbial community evenness and function in slow sand filters. Mbio 6:e00729-e815. https://doi.org/10.1128/mBio.00729-15
Haig SJ, Schirmer M, D’Amore R, Gibbs J, Davies RL, Collins G, Quince C (2015b) Stable-isotope probing and metagenomics reveal predation by protozoa drives E. coli removal in slow sand filters. ISME J 9:797–808. https://doi.org/10.1038/ismej.2014.175
Hammes F, Berney M, Wang Y, Vital M, Köster O, Egli T (2008) Flow-cytometric total bacterial cell counts as a descriptive microbiological parameter for drinking water treatment processes. Water Res 42:269–277. https://doi.org/10.1016/j.watres.2007.07.009
Hammes F, Velten S, Egli T, Juhna T (2011) Biotreatment of drinking water. In: Moo-Young M, Butler M, Webb C, Moreira A, Grodzinski B, Cui ZF, Agathos S (eds) Comprehensive biotechnology, 2nd edn. Elsevier, pp 517–530
Herrero M, Stuckey DC (2015) Bioaugmentation and its application in wastewater treatment: a review. Chemosphere 140:119–128. https://doi.org/10.1016/j.chemosphere.2014.10.033
Hijnen WAM, Dullemont YJ, Schijven JF, Hanzens-Brouwer AJ, Rosielle M, Medema G (2007) Removal and fate of Cryptosporidium parvum, Clostridium perfringens and small-sized centric diatoms (Stephanodiscus hantzschii) in slow sand filters. Water Res 41:2151–2162. https://doi.org/10.1016/j.watres.2007.01.056
Hijnen WAM, Medema GJ (2010) Elimination of micro-organisms by drinking water treatment processes: a review. IWA Publishing. https://doi.org/10.2166/9781780401584
Hijnen WAM, Schijven JF, Bonné P, Visser A, Medema GJ (2004) Elimination of viruses, bacteria and protozoan oocysts by slow sand filtration. Water Sci Technol 50:147–154. https://doi.org/10.2166/wst.2004.0044
Hijnen WAM, Suylen GMH, Bahlman JA, Brouwer-Hanzens A, Medema GJ (2010) GAC adsorption filters as barriers for viruses, bacteria and protozoan (oo)cysts in water treatment. Water Res 44:1224–1234. https://doi.org/10.1016/j.watres.2009.10.011
Horemans B, Raes B, Vandermaesen J, Simanjuntak Y, Brocatus H, T’Syen J, Degryse J, Boonen J, Wittebol J, Lapanje A, Sørensen SR, Springael D (2017) Biocarriers improve bioaugmentation efficiency of a rapid sand filter for the treatment of 2,6-dichlorobenzamide-contaminated drinking water. Environ Sci Technol 51:1616–1625. https://doi.org/10.1021/acs.est.6b05027
Inkinen J, Jayaprakash B, Santo Domingo JW, Keinänen-Toivola MM, Ryu H, Pitkänen T (2016) Diversity of ribosomal 16S DNA- and RNA-based bacterial community in an office building drinking water system. J Appl Microbiol 120:1723–1738. https://doi.org/10.1111/jam.13144
Inkinen J, Jayaprakash B, Siponen S, Hokajärvi AM, Pursiainen A, Ikonen J, Ryzhikov I, Täubel M, Kauppinen A, Paananen J, Miettinen IT, Torvinen E, Kolehmainen M, Pitkänen T (2019) Active eukaryotes in drinking water distribution systems of ground and surface waterworks. Microbiome 7:99. https://doi.org/10.1186/s40168-019-0715-5
Jehmlich N, Vogt C, Lünsmann V, Richnow HH, von Bergen M (2016) Protein-SIP in environmental studies. Curr Opin Biotechnol 41:26–33. https://doi.org/10.1016/j.copbio.2016.04.010
Jia L, Sun H, Zhou Q, Dai R, Wu W (2022) Integrated evaluation for advanced removal of nitrate and phosphorus in novel PHBV/ZVI-based biofilters: insight into functional genes and key enzymes. J Clean Prod 349:131199. https://doi.org/10.1016/j.jclepro.2022.131199
Koo H, Hakim JA, Morrow CD, Eipers PG, Davila A, Andersen DT, Bej AK (2017) Comparison of two bioinformatics tools used to characterize the microbial diversity and predictive functional attributes of microbial mats from Lake Obersee. J Microbiol Methods 140:15–22. https://doi.org/10.1016/j.mimet.2017.06.017
Kirisits MJ, Emelko MB, Pinto AJ (2019) Applying biotechnology for drinking water biofiltration: advancing science and practice. Curr Opin Biotechnol 57:197–204. https://doi.org/10.1016/j.copbio.2019.05.009
Kleiner M, Dong X, Hinzke T, Wippler J, Thorson E, Mayer B, Strous M (2018) Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities. Proc Natl Acad Sci USA 115:E5576–E5584. https://doi.org/10.1073/pnas.1722325115
Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, Li C, Strous M (2017) Assessing species biomass contributions in microbial communities via metaproteomics. Nat Commun 8:1558. https://doi.org/10.1038/s41467-017-01544-x
Knezev A (2015) Microbial activity in granular activated carbon filters in drinking water treatment PhD thesis. Wageningen University, Wageningen. https://library.wur.nl/WebQuery/wda/2081182
Kumar A, Ng DHP, Wu Y, Cao B (2019) Microbial community composition and putative biogeochemical functions in the sediment and water of tropical granite quarry lakes. Microb Ecol 77:1–11. https://doi.org/10.1007/s00248-018-1204-2
Kyselková M, Salles JF, Dumestre A, Benoit Y, Grundmann GL (2019) Distinct bacterial consortia established in ETBE-degrading enrichments from a polluted aquifer. Appl Sci 9:4247. https://doi.org/10.3390/app9204247
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/NBT.2676
Lautenschlager K, Hwang C, Ling F, Liu WT, Boon N, Köster O, Egli T, Hammes F (2014) Abundance and composition of indigenous bacterial communities in a multi-step biofiltration-based drinking water treatment plant. Water Res 62:40–52. https://doi.org/10.1016/j.watres.2014.05.035
Lee CO, Boe-Hansen R, Musovic S, Smets B, Albrechtsen HJ, Binning P (2014) Effects of dynamic operating conditions on nitrification in biological rapid sand filters for drinking water treatment. Water Res 64:226–236. https://doi.org/10.1016/j.watres.2014.07.001
Li Q, Yu S, Li L, Liu G, Gu Z, Liu M, Liu Z, Ye Y, Xia Q, Ren L (2017) Microbial communities shaped by treatment processes in a drinking water treatment plant and their contribution and threat to drinking water safety. Front Microbiol 8:2465. https://doi.org/10.3389/fmicb.2017.02465
Li XK, Chu ZR, Liu YJ, Zhu MT, Yang L, Zhang J (2013) Molecular characterization of microbial populations in full-scale biofilters treating iron, manganese and ammonia containing groundwater in Harbin, China. Bioresour Technol 147:234–239. https://doi.org/10.1016/j.biortech.2013.08.008
Li X, Upadhyaya G, Yuen W, Brown J, Morgenroth E, Raskin L (2010) Changes in the structure and function of microbial communities in drinking water treatment bioreactors upon addition of phosphorus. Appl Environ Microbiol 76:7473–7481. https://doi.org/10.1128/AEM.01232-10
Liao X, Chen C, Wang Z, Wan R, Chang CH, Zhang X, Xie S (2013a) Changes of biomass and bacterial communities in biological activated carbon filters for drinking water treatment. Process Biochem 48(2):312–316. https://doi.org/10.1016/j.procbio.2012.12.016
Liao X, Chen C, Wang Z, Wan R, Chang CH, Zhang X, Xie S (2013b) Pyrosequencing analysis of bacterial communities in drinking water biofilters receiving influents of different types. Process Biochem 48(4):703–707. https://doi.org/10.1016/j.procbio.2013.02.033
Liao X, Chen C, Zhang J, Dai Y, Zhang X, Xie S (2015) Operational performance, biomass and microbial community structure: impacts of backwashing on drinking water biofilter. Environ Sci Pollut Res Int 22(1):546–554. https://doi.org/10.1007/s11356-014-3393-7
Lin X, McKinley J, Resch CT, Kaluzny R, Lauber CL, Fredrickson J, Knight R, Konopka A (2012) Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J 6:1665–1676. https://doi.org/10.1038/ismej.2012.26
Liu C, Olivares CI, Pinto AJ, Lauderdale CV, Brown J, Selbes M, Karanfil T (2017) The control of disinfection byproducts and their precursors in biologically active filtration processes. Water Res 124:630–653. https://doi.org/10.1016/j.watres.2017.07.080
Lourenço N, Nunes LM (2017) Optimization of a vermifiltration process for treating urban wastewater. Ecol Eng 100:138–146. https://doi.org/10.1016/j.ecoleng.2016.11.074
Lueders T, Dumont MG, Bradford L, Manefield M (2016) RNA-stable isotope probing: from carbon flow within key microbiota to targeted transcriptomes. Curr Opin Biotechnol 41:83–89. https://doi.org/10.1016/j.copbio.2016.05.001
Ma B, LaPara TM, Hozalski RM (2020) The microbiome of drinking water biofilters is influenced by environmental factors and engineering decisions but has little influence on the microbiome of the filtrate. Environ Sci Technol 54:11526–11535. https://doi.org/10.1021/acs.est.0c01730
Madigan M, Martinko J, Dunlap P, Clark D (2008) Brock biology of microorganisms 12th edn. Int Microbiol 11:65-73. https://doi.org/10.2436/im.v11i1.9650
Magic-Knezev A, Wullings B, Van Der Kooij D (2008) Polaromonas and Hydrogenophaga species are the predominant bacteria cultured from granular activated carbon filters in water treatment. J Appl Microbiol 107:1457–1467. https://doi.org/10.1111/j.1365-2672.2009.04337.x
Marcantonio C, Bertelkamp C, Van BN, Pronk TE, Peer HA, Van Der WP, Brunner AM, Di Marcantonio C, Bertelkamp C, van Bel N, Pronk TE, Timmers PHA, van der Wielen P, Brunner AM (2020) Organic micropollutant removal in full-scale rapid sand filters used for drinking water treatment in the Netherlands and Belgium. Chemosphere 260:127630. https://doi.org/10.1016/j.chemosphere.2020.127630
Mattes TE, Alexander AK, Richardson PM, Munk AC, Han CS, Stothard P, Coleman NV (2008) The genome of Polaromonas sp. strain JS666: Insights into the evolution of a hydrocarbon- and xenobiotic-degrading bacterium, and features of relevance to biotechnology. Appl Environ Microbiol 74:6405–6416. https://doi.org/10.1128/AEM.00197-08
Matuzahroh N, Fitriani N, Ardiyanti PE, Kuncoro EP, Budiyanto WD, Isnadina DRM, Wahyudianto FE, Radin Mohamed RMS (2020) Behavior of schmutzdecke with varied filtration rates of slow sand filter to remove total coliforms. Heliyon 6:e03736. https://doi.org/10.1016/j.heliyon.2020.e03736
Maurya A, Singh MK, Kumar S (2020) Biofiltration technique for removal of waterborne pathogens. Waterborne Pathogens. https://doi.org/10.1016/b978-0-12-818783-8.00007-4
McDonald D, Price MN, Goodrich J, Nawrocki EP, Desantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. https://doi.org/10.1038/ismej.2011.139
Moreira NA, Bondelind M (2017) Safe drinking water and waterborne outbreaks. J Water Health 15:83–96. https://doi.org/10.2166/wh.2016.103
Murdoch RW, Hay AG (2015) The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu-1. Biodegradation 26:105–113. https://doi.org/10.1007/s10532-015-9719-4
Musat N, Musat F, Weber PK, Pett-Ridge J (2016) Tracking microbial interactions with NanoSIMS. Curr Opin Biotechnol 41:114–121. https://doi.org/10.1016/j.copbio.2016.06.007
Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S (2011) A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. https://doi.org/10.1128/mmbr.00028-10
Nyberg ET, White SA, Jeffers SN, Bridges WC (2014) Removal of plant pathogen propagules from irrigation runoff using slow filtration systems: quantifying physical and biological components. Water Air Soil Pollut 225:1–11. https://doi.org/10.1007/s11270-014-1999-5
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. https://doi.org/10.1093/nar/gkv1189
Oh S, Hammes F, Liu WT (2018) Metagenomic characterization of biofilter microbial communities in a full-scale drinking water treatment plant. Water Res 128:278–285. https://doi.org/10.1016/j.watres.2017.10.054
Palansooriya KN, Yang Y, Tsang YF, Sarkar B, Hou D, Cao X, Meers E, Rinklebe J, Kim KH, Ok YS (2020) Occurrence of contaminants in drinking water sources and the potential of biochar for water quality improvement: a review. Crit Rev Environ Sci Technol 50:549–611. https://doi.org/10.1080/10643389.2019.1629803
Palomo A, Jane Fowler S, Gülay A, Rasmussen S, Sicheritz-Ponten T, Smets BF, Fowler SJ, Gülay A, Rasmussen S, Sicheritz-Ponten T, Smets BF (2016) Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J 10:2569–2581. https://doi.org/10.1038/ismej.2016.63
Pandey PK, Kass PH, Soupir ML, Biswas S, Singh VP (2014) Contamination of water resources by pathogenic bacteria. AMB Express 4:1–16. https://doi.org/10.1186/s13568-014-0051-x
Pfannes KR, Langenbach KMWW, Pilloni G, Stührmann T, Euringer K, Lueders T, Neu TR, Müller JA, Kästner M, Meckenstock RU (2015) Selective elimination of bacterial faecal indicators in the Schmutzdecke of slow sand filtration columns. Appl Microbiol Biotechnol 99:10323–10332. https://doi.org/10.1007/s00253-015-6882-9
Pick FC, Fish KE, Biggs CA, Moses JP, Moore G, Boxall JB (2019) Application of enhanced assimilable organic carbon method across operational drinking water systems. PLoS ONE 14:1–24. https://doi.org/10.1371/journal.pone.0225477
Poghosyan L, Koch H, Frank J, van Kessel MA, Cremers G, van Alen T, Jetten MS, den Camp HJ, Lücker S (2020) Metagenomic profiling of ammonia- and methane-oxidizing microorganisms in a Dutch drinking water treatment plant. bioRxiv. https://doi.org/10.1101/2020.05.19.103440
Pontius FW (2003) Update on USEPA’s drinking water regulations. J Am Water Works Assoc 95:8–12. https://doi.org/10.1002/J.1551-8833.2003.TB10314.X
Prevost M, Laurent P, Servais P, Joret JC (2005) Biodegradable organic matter in drinking water treatment and distribution. American Water Works, Denver
Prosser JI (2020) Putting science back into microbial ecology: a question of approach. Philos Trans R Soc B Biol Sci 375:20190240. https://doi.org/10.1098/rstb.2019.0240
Qi W, Li W, Zhang J, Wu X, Zhang J, Zhang W (2019) Effect of biological activated carbon filter depth and backwashing process on transformation of biofilm community. Front Environ Sci Eng 13:1–11. https://doi.org/10.1007/s11783-019-1100-0
Ramond JB, Welz PJ, Tuffin MI, Burton SG, Cowan DA (2013) Assessment of temporal and spatial evolution of bacterial communities in a biological sand filter mesocosm treating winery wastewater. J Appl Microbiol 115:91–101. https://doi.org/10.1111/jam.12203
Schijven JF, Van den Berg HHJL, Colin M, Dullemont Y, Hijnen WAM, Magic-Knezev A, Oorthuizen WA, Wubbels G (2013) A mathematical model for removal of human pathogenic viruses and bacteria by slow sand filtration under variable operational conditions. Water Res 47:2592–2602. https://doi.org/10.1016/j.watres.2013.02.027
Shannon MA, Bohn PW, Elimelech M, Georgiadis JG, Marĩas BJ, Mayes AM (2008) Science and technology for water purification in the coming decades. Nature 452:301–310. https://doi.org/10.1038/nature06599
Simpson DR (2008) Biofilm processes in biologically active carbon water purification. Water Res 42:2839–2848. https://doi.org/10.1016/j.watres.2008.02.025
Singer E, Wagner M, Woyke T (2017) Capturing the genetic makeup of the active microbiome in situ. ISME J 11:1949–1963. https://doi.org/10.1038/ismej.2017.59
Stolz A, Schmidt-Maag C, Denner EBM, Busse HJ, Egli T, Kämpfer P (2000) Description of Sphingomonas xenophaga sp. nov. for strains BN6T and N, N which degrade xenobiotic aromatic compounds. Int J Syst Evol Microbiol 50:35–41. https://doi.org/10.1099/00207713-50-1-35
Stott R, May E, Matsushita E, Warren A (2001) Protozoan predation as a mechanism for the removal of cryptosporidium oocysts from wastewaters in constructed wetlands. Water Sci Technol 44:191–198. https://doi.org/10.2166/wst.2001.0828
Tatari K, Musovic S, Gülay A, Dechesne A, Albrechtsen HJ, Smets BF (2017) Density and distribution of nitrifying guilds in rapid sand filters for drinking water production: dominance of Nitrospira spp. Water Res 127:239–248. https://doi.org/10.1016/j.watres.2017.10.023
Terry LG, Summers RS (2018) Biodegradable organic matter and rapid-rate biofilter performance: a review. Water Res 128:234–245. https://doi.org/10.1016/j.watres.2017.09.048
Unger M, Collins MR (2008) Assessing Escherichia coli removal in the schmutzdecke of slow-rate biofilters. J Am Water Work Assoc 100:60–73. https://doi.org/10.1002/j.1551-8833.2008.tb09799.x
Velten S, Boller M, Köster O, Helbing J, Weilenmann HU, Hammes F (2011) Development of biomass in a drinking water granular active carbon (GAC) filter. Water Res 45:6347–6354. https://doi.org/10.1016/j.watres.2011.09.017
Verma S, Daverey A, Sharma A (2017) Slow sand filtration for water and wastewater treatment–a review. Environ Technol Rev 6:47–58. https://doi.org/10.1080/21622515.2016.1278278
Vignola M, Werner D, Wade MJ, Meynet P, Davenport RJ (2018) Medium shapes the microbial community of water filters with implications for effluent quality. Water Res 129:499–508. https://doi.org/10.1016/j.watres.2017.09.042
Wakelin S, Page D, Dillon P, Pavelic P, Abell GCJ, Gregg AL, Brodie E, DeSantis TZ, Goldfarb KC, Anderson G (2011) Microbial community structure of a slow sand filter Schmutzdecke: a phylogenetic snapshot based on rRNA sequence analysis. Water Sci Technol Water Supply 11:426–436. https://doi.org/10.2166/ws.2011.063
Wakelin SA, Page DW, Pavelic P, Gregg AL, Dillon PJ (2010) Rich microbial communities inhabit water treatment biofilters and are differentially affected by filter type and sampling depth. Water Sci Technol Water Supply 10:145–156. https://doi.org/10.2166/ws.2010.570
Wang H, Pryor MA, Edwards MA, Falkinham JO, Pruden A (2013) Effect of GAC pre-treatment and disinfectant on microbial community structure and opportunistic pathogen occurrence. Water Res 47:5760–5772. https://doi.org/10.1016/j.watres.2013.06.052
Wang J, de Ridder D, van der Wal A, Sutton NB (2020) Harnessing biodegradation potential of rapid sand filtration for organic micropollutant removal from drinking water: a review. Crit Rev Environ Sci Technol 51:2086–2118. https://doi.org/10.1080/10643389.2020.1771888
Weber-Shirk ML, Dick RI (1997) Biological mechanisms in slow sand filters. J Am Water Works Assoc 89:72–83
Wemheuer F, Taylor JA, Daniel R, Johnston E, Meinicke P, Thomas T, Wemheuer B (2020) Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiomes 15:1–12. https://doi.org/10.1186/s40793-020-00358-7
World Health Organization (2017) Guidelines for drinking-water quality: fourth edition incorporating the first addendum. Geneva. https://www.who.int/publications/i/item/9789241549950
Wu T, Fu GY, Sabula M, Brown T (2014) Bacterial community in the biofilm of granular activated carbon (GAC) PreBiofilter in bench-scale pilot plants for surface water pretreatment. World J Microbiol Biotechnol 30:3251–3262. https://doi.org/10.1007/s11274-014-1752-7
Yogafanny E, Fuchs S, Obst U (2014) Study of slow sand filtration in removing total coliforms and E. coli. J Sains Teknologi Lingkung 6:107–116. https://doi.org/10.20885/jstl.vol6.iss2.art4
Yusuf A, O’Flynn D, White B, Holland L, Parle-Mcdermott A, Lawler J, McCloughlin T, Harold D, Huerta B, Regan F (2021) Monitoring of emerging contaminants of concern in the aquatic environment: a review of studies showing the application of effect-based measures. Anal Methods 13:5120–5143. https://doi.org/10.1039/d1ay01184g
Zhai J, Wang Z, Shi P, Long C (2017) Microbial community in a biofilter for removal of low load nitrobenzene waste gas. PLoS ONE 12:e0170417. https://doi.org/10.1371/journal.pone.0170417
Zhang S, Gitungo SW, Axe L, Raczko RF, Dyksen JE (2017) Biologically active filters – an advanced water treatment process for contaminants of emerging concern. Water Res 114:31–41. https://doi.org/10.1016/j.watres.2017.02.014
Zhao X, Yang Y, Feng K, Wang X, Liu B, Xie G, Xing D (2021) Self-regulating microbiome networks ensure functional resilience of biofilms in sand biofilters during manganese load fluctuations. Water Res 188:116473. https://doi.org/10.1016/j.watres.2020.116473
Zhou Z, Xu L, Zhu L, Liu Y, Shuai X, Lin Z, Chen H (2021) Metagenomic analysis of microbiota and antibiotic resistome in household activated carbon drinking water purifiers. Environ Int 148:106394. https://doi.org/10.1016/j.envint.2021.106394
Yachi S, Loreau M (1999) Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci USA 96(4):1463–8. https://doi.org/10.1073/pnas.96.4.1463
Funding
This work is performed within the NWO-Dunea-Vitens partnership programme. The programme is funded by NWO, the national research council of the Netherlands (Project No. 17840).
Author information
Authors and Affiliations
Contributions
XB, GM and ID conceived this review. XB performed the literature search and data analysis and wrote the original manuscript draft. GM and ID critically revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies involving human or animal participants conducted by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bai, X., Dinkla, I.J.T. & Muyzer, G. Microbial ecology of biofiltration used for producing safe drinking water. Appl Microbiol Biotechnol 106, 4813–4829 (2022). https://doi.org/10.1007/s00253-022-12013-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12013-x