Abstract
Catalases and peroxidases are the most important enzymes that degrade hydrogen peroxide into water and oxygen. These enzymes and superoxide dismutase are the first lines of cell defense against reactive oxygen species. Metarhizium anisopliae displays an increase in catalase–peroxidase activity during germination and growth. To determine the importance of catalase during the invasion process of M. anisopliae, we isolated the cat1 gene. cat1 cDNA expression in Escherichia coli and the subsequent purification of the protein confirmed that the cat1 gene codes for a monofunctional catalase. Expression analysis of this gene by RT-PCR from RNA isolated from fungus grown in liquid cultures showed a decrease in the expression level of the cat1 gene during germination and an increase during mycelium growth. The expression of this gene in the fungus during the infection process of the larvae of Plutella xylostella also showed a significant increase during invasive growth. Transgenic strains overexpressing the cat1 gene had twice the catalase activity of the wild-type strain. This increase in catalase activity was accompanied by a higher level of resistance to exogenous hydrogen peroxide and a reduction in the germination time. This improvement was also observed during the infection of P. xylostella larvae. M. anisopliae transgenic strains overexpressing the cat1 gene grew and spread faster in the soft tissue of the insect, reducing the time to death of the insect by 25% and the dose required to kill 50% of the population 14-fold.





Similar content being viewed by others
References
Abbott DA, Suir E, Duong GH, de Hulster E, Pronk JT, van Maris AJ (2009) Catalase overexpression reduces lactic acid-induced oxidative stress in Saccharomyces cerevisiae. Appl Environ Microbiol 75:2320–2325
Braga GU, Destefano RH, Messias CL (1999) Oxygen consumption by Metarhizium anisopliae during germination and growth on different carbon sources. J Invertebr Pathol 74:112–119
Brown-Peterson NJ, Salin ML (1993) Purification of a catalase-peroxidase from Halobacterium halobium: characterization of some unique properties of the halophilic enzyme. J Bacteriol 175:4197–4202
Crespo R, Pedrini N, Juarez MP, Dal Bello GM (2008) Volatile organic compounds released by the entomopathogenic fungus Beauveria bassiana. Microbiol Res 163:148–151
Dutra V, Nakazato L, Broetto L, Silveira Schrank I, Henning Vainstein M, Schrank A (2004) Application of representational difference analysis to identify sequence tags expressed by Metarhizium anisopliae during the infection process of the tick Boophilus microplus cuticle. Res Microbiol 155:245–251
Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH (1998) Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol Microbiol 27:121–130
Freimoser FM, Screen S, Bagga S, Hu G, St Leger RJ (2003) Expressed sequence tag (EST) analysis of two subspecies of Metarhizium anisopliae reveals a plethora of secreted proteins with potential activity in insect hosts. Microbiology 149:239–247
Freimoser FM, Hu G, St Leger RJ (2005) Variation in gene expression patterns as the insect pathogen Metarhizium anisopliae adapts to different host cuticles or nutrient deprivation in vitro. Microbiology 151:361–371
Garre V, Tenberge KB, Eising R (1998) Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: putative role in pathogenicity and suppression of host defense. Phytopathology 88:744–753
Giles SS, Stajich JE, Nichols C, Gerrald QD, Alspaugh JA, Dietrich F, Perfect JR (2006) The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell 5:1447–1459
Goettel MS, Leger RJ, Bhaire S, Jung MK, Oakley BR, Roberts D, Staples RC (1990) Pathogenicity and growth of Metarhizium anisopliae stably transformed to benomyl resistance. Curr Genet 17:129–132
Hisada H, Hata Y, Kawato A, Abe Y, Akita O (2005) Cloning and expression analysis of two catalase genes from Aspergillus oryzae. J Biosci Bioeng 99:562–568
Kawasaki L, Aguirre J (2001) Multiple catalase genes are differentially regulated in Aspergillus nidulans. J Bacteriol 183:1434–1440
Kawasaki L, Wysong D, Diamond R, Aguirre J (1997) Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J Bacteriol 179:3284–3292
Lledias F, Rangel P, Hansberg W (1998) Oxidation of catalase by singlet oxygen. J Biol Chem 273:10630–10637
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Michan S, Lledias F, Hansberg W (2003) Asexual development is increased in Neurospora crassa cat-3-null mutant strains. Eukaryot Cell 2:798–808
Miller CD, Rangel D, Braga GU, Flint S, Kwon SI, Messias CL, Roberts DW, Anderson AJ (2004) Enzyme activities associated with oxidative stress in Metarhizium anisopliae during germination, mycelial growth, and conidiation and in response to near-UV irradiation. Can J Microbiol 50:41–49
Mutoh N, Nakagawa CW, Yamada K (1999) The role of catalase in hydrogen peroxide resistance in fission yeast Schizosaccharomyces pombe. Can J Microbiol 45:125–129
Nakazato L, Dutra V, Broetto L, Staats CC, Vainstein MH, Schrank A (2006) Development of an expression vector for Metarhizium anisopliae based on the tef-1α homologous promoter. Appl Microbiol Biotechnol 72:521–528
Paris S, Wysong D, Debeaupuis JP, Shibuya K, Philippe B, Diamond RD, Latge JP (2003) Catalases of Aspergillus fumigatus. Infect Immun 71:3551–3562
Pedrini N, Juarez MP, Crespo R, de Alaniz MJ (2006) Clues on the role of Beauveria bassiana catalases in alkane degradation events. Mycologia 98:528–534
Roberts DW, St Leger RJ (2004) Metarhizium spp., cosmopolitan insect-pathogenic fungi: mycological aspects. Adv Appl Microbiol 54:1–70
Sambrook JG, Russell R (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schliebs W, Wurtz C, Kunau WH, Veenhuis M, Rottensteiner H (2006) A eukaryote without catalase-containing microbodies: Neurospora crassa exhibits a unique cellular distribution of its four catalases. Eukaryot Cell 5:1490–1502
Shima S, Netrusov A, Sordel M, Wicke M, Hartmann GC, Thauer RK (1999) Purification, characterization, and primary structure of a monofunctional catalase from Methanosarcina barkeri. Arch Microbiol 171:317–323
Skamnioti P, Henderson C, Zhang Z, Robinson Z, Gurr SJ (2007) A novel role for catalase B in the maintenance of fungal cell-wall integrity during host invasion in the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 20:568–580
St Leger R, Cooper R, Charnley A (1986) Cuticle-degrading enzymes of entomopathogenic fungi: regulation of production of chitinolytic enzymes. J Gen Microbiol 132:1509–1517
St Leger RJ, Bidochka MJ, Roberts DW (1994) Isoforms of the cuticle-degrading Pr1 proteinase and production of a metalloproteinase by Metarhizium anisopliae. Arch Biochem Biophys 313:1–7
St Leger R, Joshi L, Bidochka MJ, Roberts DW (1996) Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc Natl Acad Sci USA 93:6349–6354
St Leger RJ, Joshi L, Roberts DW (1997) Adaptation of proteases and carbohydrates of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 143(Pt 6):1983–1992
Staats CC, Junges A, Fitarelli M, Furlaneto MC, Vainstein MH, Schrank A (2007) Gene inactivation mediated by Agrobacterium tumefaciens in the filamentous fungi Metarhizium anisopliae. Appl Microbiol Biotechnol 76:945–950
Wang C, St Leger RJ (2005) Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot Cell 4:937–947
Wang N, Yoshida Y, Hasunuma K (2007) Loss of Catalase-1 (Cat-1) results in decreased conidial viability enhanced by exposure to light in Neurospora crassa. Mol Genet Genomics 277:13–22
Wayne LG, Diaz GA (1986) A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal Biochem 157:89–92
Yamashita K, Shiozawa A, Banno S, Fukumori F, Ichiishi A, Kimura M, Fujimura M (2007) Involvement of OS-2 MAP kinase in regulation of the large-subunit catalases CAT-1 and CAT-3 in Neurospora crassa. Genes Genet Syst 82:301–310
Zamocky M, Furtmüller PG, Obinger C (2008) Evolution of catalases from bacteria to humans. Antioxid Redox Signal 10:1527–1548
Acknowledgments
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT), Consejo Estatal de Ciencia y Tecnología del Estado de Guanajuato (CONCyTEG), Secretaria de Educacion Publica (SEP), and the University of Guanajuato. CEMH and IEPG were recipients of a fellowship from CONACyT, México.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Fig. S1
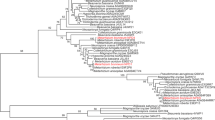
Comparison of Metarhizium anisopliae CAT1 with the most similar monofunctional catalases and catalase/peroxidases. M. anisopliae, Macat1* was aligned with catalases from: Aspergillus fumigatus, Af: catA (accession number P78574), catB (Q92405); A. nidulans, An: catA (P55305), catB (P78619), katG (Q96VT4); A. niger, Ani: catA (A2QT57), katG (A2Q7T1); A. oryzae, Ao: catA (Q877A1), catB (Q877A8), katG (Q2TW34); Botrytis cinerea, Bc: cat2 (G96WN1), catA (P55304); Candida albicans, Ca: CTA1 (O13289); Caenorhabditis elegans, Ce: ctl-1 (O61235), ctl-2 (Q27487), ctl-3 (Q8MYL7); Cladosporium fulvum, Cf: cat-2 (Q9C476); Cochliobolus heterostrophus, Ch: CAT2 (Q6UJ33); Claviceps purpurea, Cp: cat1 (O60038); Escherichia coli, Ec: katE (P21179), katG (P13029); Gibberella moniliformis, Gm: cat2 (B8XX03); Magnaporte grisea, Mg: katG1 (A4R559), MGG_10061 (A4R6C5), MGG_06442 (A4R8L1); Neurospora crassa, Nc cat-1: (Q9C168), cat-2 (Q8X182), cat-3 (Q9C169); Podospora anserina, Pa: catA (Q9HDP6), catB (Q9HDD5); Paracoccidioides brasiliensis, Pb: catA (Q6RSH8); Penicillium marneffei, Pm: katG (Q8NJN2); Saccharomyces cerevisiae, Sc: CTA1 (P15202), CTT1 (PO6115); Sclerotinia sclerotiorum, Ss: SS1G-02784 (A7EBU8); and Ustilago maydis, Um: kat G (Q4P914) http://www.ebi.ac.uk/. a Conserved amino acid that forms part of the active site (asterisk), heme coordination (empty circle), as well as the residues involved in forming the heme pocket (filled circle) are indicated. The conserved amino acid sequence FDHERVPERAVHARGAG containing the conserved H-102 and present in all heme catalases for the proper binding and reduction of the H2O2 molecule, and the heme binding site composed of amino acid residues RIFSYLDTQL are indicated (boxes). b Phylogenetic tree of monofunctional catalases and catalase/peroxidases. The alignment was performed using the Clustal W method from Lasergene 8.0 program MegAlign (DNASTAR, Inc.) with its default parameters. (DOC 187 kb)
Fig. S2
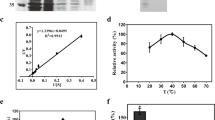
Expression and purification of recombinant catalase in E. coli. a Twenty microliters of each fraction was loaded into each lane of a 10% SDS-PAGE gel and stained with Coomassie blue. Lane 1 Crude extract from E. coli expressing CAT1 without IPTG; lane 2 Crude extract from E. coli expressing CAT1 with 1 mM IPTG induction for 3 h; lane 3 Flow-through proteins from Ni-NTA column; lane 4 Proteins eluted with 20 mM imidazole; lane 5 Proteins eluted with 40 mM imidazole; lane 6 Purified CAT1 protein eluted with 100 mM imidazole; lane M corresponds to molecular weight markers in kDa. b In-gel assay for catalase (lanes 1 and 2) or catalase and peroxidase (lanes 3 and 4). Fifty microliters of crude extract from E. coli expressing CAT1 with 1 mM IPTG induction for 3 h (lanes 1 and 3) or purified recombinant CAT1 (lane 2 and 4) was loaded into each lane of a non-denaturing 8% polyacrylamide gel, and then catalase (seen as clear zone, lanes 1 and 2) and peroxidase (seen as dark band on a clear zone, lanes 3 and 4) were visualized as described in “Materials and methods.” c In-gel assay for catalase (lane 1) or catalase and peroxidase (lane 2). Fifty micrograms of protein from cellular crude extracts of M. anisopliae Ma10 grown for 48 h in minimal medium with P. xylostella cuticle was loaded into each lane of a non-denaturing 8% polyacrylamide gel, and then catalase (lane 1) and peroxidase (lane 2) were visualized as described in “Materials and methods.” (PDF 4999 kb)
Fig. S3
In vitro expression of chit1 and cat1 from M. anisopliae. Conidia from M. anisopliae Ma10 were grown in liquid minimal media containing 2% glucose for 48 h (lane 1), liquid minimal media containing 1% chitin (lane 2) for 12 h or 1% P. xylostella cuticle (lane 3) for 12 h. The cultures were incubated in Erlenmeyer flasks at 28°C as described in “Materials and methods.” The fungal material was collected by filtration and was used to compare transcript abundance by RT-PCR analysis. a Relative RT-PCR of chit1 and AJ274118 (loading control) using total RNA. b Relative RT-PCR analysis of cat1 and AJ274118 (loading control) using total RNA (DOC 292 kb)
Fig. S4
UV/vis spectrum of purified CAT1. The UV/vis spectrum of purified CAT1 was obtained in solution with 50 μg of CAT1 in 50 mM phosphate buffer, pH 7.0. The spectrum was recorded with a spectrophotometer Ultrospec 4300 Pro (Amersham Bioscience) (DOC 79 kb)
Table S1
Primers designed for use in gene cloning, overexpression, DNA quantification, and gene expression by RT-PCR analysis (DOC 43 kb)
Rights and permissions
About this article
Cite this article
Morales Hernandez, C.E., Padilla Guerrero, I.E., Gonzalez Hernandez, G.A. et al. Catalase overexpression reduces the germination time and increases the pathogenicity of the fungus Metarhizium anisopliae . Appl Microbiol Biotechnol 87, 1033–1044 (2010). https://doi.org/10.1007/s00253-010-2517-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2517-3