Abstract
Nonylphenol (NP) is an estrogenic pollutant which is widely present in the aquatic environment. Biodegradation of NP can reduce the toxicological risk. In this study, aerobic biodegradation of NP in river sediment was investigated. The sediment used for the microcosm experiments was aged polluted with NP. The biodegradation of NP in the sediment occurred within 8 days with a lag phase of 2 days at 30°C. During the biodegradation, nitro-nonylphenol metabolites were formed, which were further degraded to unknown compounds. The attached nitro-group originated from the ammonium in the medium. Five subsequent transfers were performed from original sediment and yielded a final stable population. In this NP-degrading culture, the microorganisms possibly involved in the biotransformation of NP to nitro-nonylphenol were related to ammonium-oxidizing bacteria. Besides the degradation of NP via nitro-nonylphenol, bacteria related to phenol-degrading species, which degrade phenol via ring cleavage, are abundantly present.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nonylphenol (NP) is an estrogenic surfactant which is widely spread in the aquatic environment. Nonylphenols consist of a phenol group with a linear or a branched chain of nine carbon atoms. A technical mixture of branched nonylphenol (tNP) isomers is used for the production of nonylphenol polyethoxylates (NPEO; Wheeler et al. 1997; Thiele et al. 2004). This technical mixture does not contain linear NP (4-n-NP), and the linear form is, therefore, generally not present in natural samples. At anaerobic conditions in wastewater treatment plants, NPEO degrades partially to NP, which is a persistent end product in sewage sludges and the effluent of wastewater treatment plants (Stephanou and Giger 1982; Giger et al. 1984). When these NP-contaminated sludges and effluents are discharged, NP ends up in the environment as a pollutant.
Due to its chemical properties, NP adsorbs to sediments and accumulates in anaerobic sediments of various rivers such as the Llobregat river (Petrovic et al. 2002) and the Ebro river in Spain (Lacorte et al. 2006) and the Elbe river in the Czech Republic and Germany (Heemken et al. 2001). Nonylphenol can desorb from the sediment and re-enter the water phase where it can bioaccumulate in fishes and be harmful for other aquatic organisms (Tyler et al. 1998; Servos 1999; LeBlanc and Rider 2000; Snyder et al. 2001; Yang et al. 2005). Biodegradation of NP can reduce the toxicological risk of NP in the environment.
Under aerobic conditions, bacterial degradation of NP (branched and linear) has frequently been shown (Ekelund et al. 1993; Hesselsoe et al. 2001; Corvini et al. 2006a). However, anaerobic biodegradation of branched NP by bacteria is not observed so far. Only anaerobic biodegradation of linear NP has been described to occur in sediment from the Erren river in China under sulfate-reducing, nitrate-reducing, and methanogenic conditions (Chang et al. 2004). Aerobic NP-degrading bacterial strains are isolated from various environments. For example, Sphingomonas sp. TTNP3 and Sphingobium xenophagum Bayram, formerly known as Sphingomonas xenophaga Bayram, are isolated from wastewater treatment plants (Tanghe et al. 1999; Gabriel et al. 2005a). Stenotrophomonas sp. and Pseudomonas mandelii are isolated from polluted soils (Soares et al. 2003) and Sphingobium amiense, formerly known as Sphingomonas sp. YT (de Vries et al. 2001; Ushiba et al. 2003), from sediment. Most isolates are able to use NP as a sole carbon and energy source (Tanghe et al. 1999; Soares et al. 2003; Gabriel et al. 2005a), whereas some strains can only degrade NP cometabolically in the presence of yeast extract as carbon and energy source like S. amiense (de Vries et al. 2001).
Three possible bacterial degradation pathways of NP are mentioned in literature. One suggested pathway is the degradation of NP via a nonylphenol metabolite with a nitro-group at the ortho-position of the aromatic ring (Fig. 1). This metabolite was observed in the degradation experiments with soil and sewage sludge (Telscher et al. 2005) and in agricultural soils (Zhang et al. 2009). A second postulated pathway starts with the cleavage of the phenolic ring via the phenolic degradation pathway. This pathway is suggested for the biodegradation of NP by Pseudomonas spp., which was isolated from contaminated soil from a former industrial site (Soares et al. 2003). A third pathway of NP degradation starts with the hydroxylation of the C-4 atom of the aromatic ring to which the nonyl chain is attached, also called ipso-hydroxylation. This pathway is intensively studied in S. xenophagum Bayram and Sphingomonas sp. TTNP3. These strains were isolated from activated sludge from a municipal wastewater treatment plant and a lab-scale activated sludge reactor fed with NP, respectively (Tanghe et al. 1999; Gabriel et al. 2005a, b; Corvini et al. 2006b, 2007; Kohler et al. 2008).
This paper describes the aerobic biodegradation of the technical mixture of branched NP in NP-polluted river sediment. During the biodegradation process, the formation of nitro-nonylphenol was observed. The origin of the nitro-group was studied by using medium containing labeled ammonium nitrate (15NH4NO3 or NH 154 NO3), and the possible involved microorganisms were identified by culture-independent approaches.
Materials and methods
Chemicals and stock solutions
Nonylphenol technical mixture (purity >94%) and linear NP (pestanal, purity >99%) were purchased from Riedel de Haën (Seelze, Germany). Labeled 15NH4NO3 (98% labeled) was ordered from Sigma-Aldrich (Zwijndrecht, The Netherlands) and NH 154 NO3 (98% labeled) from Isotech (Miamisburg, OH, USA). Solvents and chemicals were of the highest purity available and were used without further purification.
For the addition of tNP to the degradation experiments (22 μM of final concentration), a highly concentrated suspension of tNP (3.5 mM) in water was prepared. UV-sterilized tNP was used. During the UV-sterilization, no photo oxidized products or metabolites were formed as confirmed by gas chromatography-mass spectrometry (GC-MS) analyses. Before using the suspension, the bottle with the suspension was heated at 60°C and shaken to homogenize the suspension. Homogeneity of the suspension was tested by analyzing four sets of triplicates by GC-MS with solid-phase micro extraction (SPME), reaching 24% of standard deviation. By using this suspension, it was possible to add and re-add tNP as single carbon source in a small volume to batch experiments in a concentration of 22 μM without the addition of an unwanted solvent, which may influence the biodegradation process.
Biodegradation experiments of tNP in slurries with originally polluted sediments
Nonylphenol polluted sediment was used for the biodegradation experiments and was collected in June 2005 from the Huerva river in Zaragoza (Spain; 41°37′23″ N, 0°54′28″ W), a tributary of the Ebro river. During the sampling, the temperature of the river water was 25.1°C. Sediment was taken with stainless steel cores to 50 cm depth and transported on ice to the laboratory. In the laboratory, the sediment was sieved aerobically at mesh <2 mm and stored at 4°C until use. The sediment was polluted with 14 ± 1.5 mg/kg dry weight sediment of branched NP isomers. The individual branched isomers were not identified, and linear NP was not present in the polluted sediment.
The aerobic degradation of the branched NP present in the polluted sediment was studied in 250 ml serum bottles with 2 ± 0.2 g dry weight tNP-polluted sediment and 50 ml of aerobic medium, which results in 2.5 ± 0.6 nmol tNP per milliliter slurry added with the sediment. No additional tNP was added to these batches. The headspace of the bottles (200 ml) was filled with air. The aerobic medium was prepared as described in Tros et al. (1996) but without Na2SeO3·5H2O. The bottles were closed with a viton stopper with crimp cap and incubated horizontally shaken (110 rpm) in the dark at 30°C. These aerobic degradation experiments were performed in triplicate, and these batches were defined as the first generation (Scheme 1). Duplicate sterile controls were included, which were autoclaved for 1 h at 121°C, followed by addition of 50 mg/l HgCl2 to prevent biological activity during the experiment. To follow the NP degradation, slurry samples (200 μl) were taken regularly in duplicate from the batches and added to a capped 20 ml headspace vial with 1.8 ml MilliQ water with 7 mg/l HgCl2 as described previously (de Weert et al. 2008).
Transfers
Transfers of the NP-degrading cultures were made and incubated at 30°C (Scheme 1). Each transfer was considered as a new generation and was prepared with fresh aerobic medium and 10% (v/v) of inoculum of the former generation. The second-generation batches were prepared with inoculum of the sediment slurry batches.
After two transfers, a tenfold dilution series (fourth generation) was performed to further enrich the NP-degrading population. The dilution series was carried out to a maximum dilution of 1010. The 103 dilution of this fourth generation was used in further biodegradation experiments since this was the most enriched active incubation.
Transfers of this 103 dilution were made in triplicate (fifth-generation batches coded as A.5, B.5, and C.5) in fresh aerobic medium with 10% (v/v) of inoculum to determine the degradation rate of the tNP. Degradation rate constants and half-life times were calculated from fifth-generation batches by using first-order kinetics.
The sixth-generation batches (coded as A.6, B.6, and C.6) were used to investigate the degradation of the formed nitro-NP intermediates. After complete degradation of the added tNP in batch A.6 (15 days), 1 mM tNP (= 11.5 mg tNP; oversaturated) was added to this batch to study the effect of a high concentration of tNP on the microbial population.
For molecular analyses, samples from batch A.6 were taken before the addition of pure tNP (t = 15 days) and also after 17, 28, and 55 days. Samples for molecular analyses from batches B.6 and C.6 were taken after 6, 13, and 40 days of incubation.
Metabolite formation
To study the formation of the metabolites, degradation experiments were performed with tNP concentrations above the maximum water solubility. These experiments were encoded as experiment D. Batches with 2 ± 0.2 g dry weight of the original sediment polluted with a mixture of branched NP isomers and 50 ml of aerobic medium batches were prepared. The branched NP isomers in the sediment were completely degraded, after which 1 mM UV-sterile pure tNP (= 11.5 mg tNP) was added under sterile conditions. A sterile control was included, and the batches were incubated horizontally shaken (110 rpm) in the dark at 30 °C.
To be able to study the formation of an individual nitro-nonylphenol isomer, 4-n-NP was used as single NP isomer. Batches were incubated in duplicate with 4-n-NP in concentrations above the maximum water solubility (5 mg/l). Inoculum (10%, v/v) was used from the first-generation tNP-degrading slurry batch (Scheme 1).
All the batches were sampled regularly and were analyzed by GC-MS. Per analyzed sample, 200 μl slurry was taken from the bottles and added to a capped 20-ml headspace vial with 1.8 ml MilliQ water with 7 mg/l HgCl2 as described in Weert et al. (2008).
Degradation experiments of tNP with 15N-labeled NH4NO3
Inoculum from the third-generation batch (Scheme 1) was used for the degradation batches in medium with labeled 15NH4NO3 or NH 154 NO3 to study the origin of the incorporated NO2-group of the formed intermediate. Therefore, batches were prepared in triplicate with 50 ml aerobic medium with either 12.5 mM 15NH4NO3 or NH 154 NO3 and 1 mM tNP (above saturation) and 10% (v/v) of inoculum. The batches were incubated horizontally shaken (110 rpm) in the dark at 30°C. Samples were collected for GC-MS analysis at different time intervals to follow the formation of intermediates of the degradation of tNP. Individual isomers were not tested with labeled NH4NO3. Samples for molecular analyses were taken after 51 days of incubation from one of these fourth-generation batches grown with 15NH4NO3 (batch coded as D.4).
Chemical analyses
To analyze the concentration of tNP and the formation of metabolites in the batches, the collected liquid samples in the 20-ml headspace vials were measured by GC-MS with SPME injections. The NP and the metabolites were extracted with an 85-μm polyacrylate SPME fiber from the headspace of the samples and analyzed by GC-MS as described in Weert et al. (2008).
Accurate mass weight analysis was used to identify the formed metabolites with a nitro-group. The GC-MS chromatograms suggested that the incorporated NO2-group was attached to the phenolic ring and not to the chain. Therefore, the nitro-phenol metabolite from the third-generation batch with the single isomer 4-n-NP was extracted instead of the group of nitro-metabolites that were formed when tNP was used, since only one single metabolite isomer could be identified with accurate mass weight analysis. The metabolite was extracted from 15 ml medium with 10 ml dichloromethane and hexane (1:1) by horizontally shaking (110 rpm) for 2 h at 30°C. The dichloromethane/hexane was completely evaporated; the extracted metabolite was resuspended in 40 μl dichloromethane and analyzed via direct insert probe mass analysis on a Finnigan MAT95 mass spectrometer. An amount of 2 μl sample was loaded into the mass spectrometer. The analysis started at room temperature, and the temperature of the sample was increased in steps of 10°C until enough sample evaporated for analysis. Data were acquired in the electron impact mode (70 eV) at a resolution of 5,400, scanning range m/z 24–600 at 2 s per decade. Molecular ion peak m/z 265 was used for accurate mass weight analyses.
DNA extraction and polymerase chain reaction
To extract the DNA from the batches, 5 ml of a different time period of the sixth-generation batches A.6, B.6, and C.6 was filtrated through a 0.2-μm pore size cellulose acetate filter (Whatman GmbH, Dassel, Germany). Material that was attached to the viton rubber stopper from the fourth-generation batch D.4 was used as well. After filtration, DNA was extracted by using a BIO101 Systems Fast DNA®Kit for Soil (Qbiogene, Inc., CA, USA) according to the manufacturer’s instructions. Bead-beat step was modified from 30 to 45 s. The extracted DNA was stored at −20°C until further analysis.
Nested polymerase chain reaction (PCR) approach encompassing two PCR steps was performed with the extracted DNA. The first PCR on the DNA samples was performed to amplify the complete 16S rRNA genes with the bacterial primer mixture fD1/fD2 and rP1/rP2 as based on the method described by Weisburg et al. (1991). Tenfold diluted PCR amplicons of the complete 16S rRNA genes were used to carry out a clone library. Amplifications were performed with 1.25 U Takara Ex Taq DNA polymerase (Takara Bio, Otsu, Shiga, Japan).
For the denaturing gradient gel electrophoresis (DGGE), direct amplification of the DNA with the F341-GC and R534 primers was not possible. Therefore, a nested-PCR approach was performed. These nested amplifications were performed with 0.4 U Taq-polymerase (Fermentas International, Inc., Burlington, Canada), and a tenfold diluted PCR amplicon from first PCR was amplified in this nested step (second PCR reaction). The primers F341-GC, including a GC-clamp and R534, were used in the nested PCR based on the method described by Yu and Morrison (2004).
Denaturing gradient gel electrophoresis
A DGGE was performed based on the method described by Yu and Morrison (2004). Approximately 400 ng of nested-PCR product (25 μl) was loaded on an 8% (w/v) polyacrylamide gel with a denaturing gradient of 30% to 70% denaturant using a Dcode™ Universal Mutation Detection System (Bio-Rad) at 100 V at 60°C for 16 h. The gels were stained for 45 min with SybrGold (Molecular Probes, Inc., Eugene, OR, USA) in 20 ml of TAE and viewed under UV light. DGGE bands were processed using Quantity-one version 4.6.2 image analysis software (Bio-Rad Laboratories, Hercules, CA, USA) and corrected manually when needed. After normalization of the gels, bands with the relative peak area intensity above 1% were included in further analyses and were digitalized and analyzed.
Bacterial diversity, based on DGGE gel analysis, was assessed by using Shannon–Weaver diversity index (H′ = −Σ P i log P i ) (Shannon and Weaver 1949). P i is the relative peak intensity of a DGGE band, calculated as P i = n i /N, where n i is the peak area of the band and N the sum of all the peak areas in a DGGE lane. Furthermore, a covariance principal component analysis (PCA) was performed on band type and peak height with the Excel application StatistiXL, version 1.8, to consider possible shifts and similarities in microbial populations during the incubation of batches A.6, B.6, C.6, and D.4.
Cloning and sequencing
Clone libraries of 16S rRNA genes were constructed from the DNA of batch A.6 at 15 days, cultured with 15 μM tNP and of batch D.4 cultured with 1 mM tNP (saturated). The PCR amplicons of the 16S rRNA genes were purified with the QIA Quick® PCR purification kit (Qiagen, Hilden, Germany). Almost complete 16S rRNA gene fragments were cloned by using the TOPO-TA cloning Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The plasmids were purified with the Qiagen Qia prep® spin mini prep kit and sent for sequencing to Eurofins MWG Operon (Ebersberg, Germany) with M13 uni (-21) sequencing primer. Sequences were inspected for ambiguous base assignments and were compared with sequences deposited in public accessible databases using the NCBI BLAST search tool at htpp://www.ncbi.nlm.nih.gov/blast. Alignment of the sequences was done by Clustralw on line software at http://www.ebi.ac.uk/tools/clustralw2.
GenBank accession numbers
The 16S rRNA gene nucleotide sequences determined in this study have been deposited into the GenBank database under accession numbers FJ591135–FJ591150, FJ609321–FJ609385, and FJ626783–FJ626794. If sequences were identical, the longest sequence was submitted.
Results
Nonylphenol degradation
The biodegradation of the mixture of branched NP isomers that was present in the polluted sediment (first-generation batches) started after a lag phase of 2 days. Within 8 days, 95% (3.3 ± 0.4 nmol/ml slurry) of the original present NP isomers were biodegraded (data not shown). The half-life time of tNP in the sediment was 1.5 ± 0.4 day. The higher present amount of NP in the slurry compared to amount mentioned in the “Materials and methods” section is because the NP was present in the used sediment and not additionally added. The sediment was heterogeneous polluted with NP, even after making the sediment as homogenous as possible.
In the batches of the 103 diluted fifth generation, tNP was added, and a decrease in tNP concentration in batches A.5, B.5, and C.5 was observed from t = 0 (Fig. 2). The decrease during these first days was due to other processes than degradation by microorganisms, since this decrease was also observed in the sterile control. This abiotic decrease can be due to adsorption to the glassware and the viton stopper, since it is known that NP has the tendency to adsorb to surfaces (Hou et al. 2006; Navarro et al. 2009). In the fifth-generation batch A.5, the biodegradation started after 1 day, and within less than 12 days, 3 μM tNP was degraded. The biodegradation in the fifth-generation batches B.5 and C.5 started after 5 days of incubation, and 4 μM tNP was biodegraded within 10 and 11 days, respectively. The half-life time of tNP in these transfers varied from 4 to 5 days, although the data are limited for calculating the half-life times.
Metabolite formation
Addition of pure tNP at a concentration of 1 mM (above saturation) to biodegradation batches resulted in the formation of a group of metabolites as seen in the GC-MS chromatogram (Fig. 3). The metabolites were not present at the start of the experiment and were not observed in the sterile control indicating that these metabolites were formed during the degradation of NP. The peak pattern of tNP in the GC-MS chromatogram was seen between retention times 7.5 and 8.5 min (Fig. 3). The pattern consists of several peaks since tNP is a mixture of branched NP isomers. The peak pattern of the formed group of metabolites was seen between 9 and 10 min and showed the same peak pattern as the tNP. The fragmentation pattern of tNP contains ions of m/z 107, 121, 135, 149, and 191, whereas the fragmentation patterns of the metabolites showed ions of m/z 152, 166, 180, 194, and 236, indicating a mass shift of 45 m/z. The same mass shift of 45 m/z was observed in the batches growing on 4-n-NP. An estimation of the metabolite concentration was not made because the response of the group of metabolites in the GC-MS was unknown.
The similarity of the chromatogram pattern of the tNP and the group of metabolites, as well as the same mass shift of 4-n-NP, indicated the substitution of a hydrogen atom by a nitro-group at the phenolic ring (Posthumus 2008, Department of Organic Chemistry, Wageningen University, the Netherlands, personal communication). This was confirmed by the results of the accurate mass weight analysis in extract of the sample of the 4-n-NP degradation. The observed mass spectrum contained m/z 43 (11% relative intensity), 57 (9), 106 (8), 135 (11), 152 (100), and 265 (56), which was in agreement with a 2- or 3-nitro-4-nonylphenol. Nuclear magnetic resonance analysis could not be performed to identify the position of the nitro-group in the molecular structure, as the concentration of the metabolite in the extract was too low.
Incubations with tNP above the maximum solubility and labeled 15NH4NO3 or NH 154 NO3 medium were performed to investigate the origin of the nitro-group. The nitro-group attached to the NP in the batches with labeled 15NH4NO3 gave a shift in mass weight of 46 m/z, whereas the formed metabolite in the incubates with labeled NH 154 NO3 showed the earlier observed shift in mass weight of 45 m/z. The mass weight shift of 46 m/z indicated that a labeled N-atom was attached to the NP. Because the mass shift of 46 m/z was only observed with labeled ammonia, this demonstrates that the attached nitro-group in the metabolites originated from the ammonium in the medium via the formation of nitrite and not of nitrate. If the nitro-group had originated from the nitrate, a mass shift of 46 m/z would also have been observed in the batches with labeled nitrate. An abiotic reaction between NP and added nitrite (12.5 mM), resulting in the formation of nitro-nonylphenol, was not observed in the first 4 weeks of incubation. More prolonged abiotic incubation showed the formation of nitro-nonylphenol (data not shown). In our batches, the formation of nitro-nonylphenol was already observed after 10 days of incubation. This indicates that the production of nitro-NP in our batches appears to be a biological process and can also be formed in low amounts, after longer periods of incubation, through abiotic mechanisms.
Abiotic incubations with tNP above the maximum solubility and nitrite (12.5 mM) under abiotic conditions did not show the formation of nitro-nonylphenol within 4 weeks of incubation. More prolonged abiotic incubation showed the formation of nitro-nonylphenol (data not shown).
The nitro-nonylphenol metabolites accumulated in the batches with tNP concentrations above the maximum water solubility. Nitro-nonylphenol was also detected in the sixth-generation batches A.6, B.6, and C.6 cultured with only 11 μM tNP. Figure 4 shows this formation in batch A.6, before the addition of 1 mM tNP (above saturation) at t = 15, and of batch B.6. Nitro-nonylphenol was already produced during the first 10 days in batch A.6, followed by a decrease in concentration indicating that the metabolite was further degraded. After the addition of 1 mM tNP to this batch, no nitro-nonylphenol metabolites were measured in this batch. In batch B.6, nitro-nonylphenol was produced between the sixth and 12th days of incubation. At t = 13, the concentrations of tNP and the nitro-nonylphenol decreased below the detection limit (18 nM). After re-addition of 4.5 μM tNP at t = 13, an increase of nitro-nonylphenol was observed again at t = 14 followed by a decrease, again indicating the biodegradability of the formed nitro-nonylphenol. No further metabolites were observed from further degradation of the nitro-nonylphenol by our used GC-MS method.
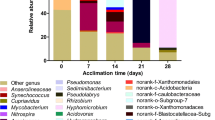
Microbial diversity
The effect of the presence of 11 μM tNP or tNP in a concentration above the maximum solubility on the microbial population was analyzed by DGGE. The band patterns in the DGGE of the four DNA samples at different times during the incubation from the sixth-generation batch A.6 contained 15 to 18 DGGE bands (Fig. 5). The Shannon–Weaver diversity index obtained from the DGGE profiles varied between 1.09 and 1.10, indicating that the diversity did not change during the incubation. Also, no change in diversity was observed after the addition of a tNP concentration above the maximum solubility at day 15, and the population remained stable. The DGGE patterns of the samples at different times of the sixth generation, batches B.6 and C.6, cultured with only 11 μM NP, contained both 16 to 18 bands and looked similar to the patterns of the samples of batch A.6. The Shannon–Weaver diversity index B.6 and C.6 ranged from 1.08 to 1.12, respectively. This indicated that the microbial diversity of batches B.6 and C.6 did not change either during the incubation of 40 days. This was confirmed by PCA (Fig. 6). In fact, the DGGE profiles of the batches A.6, B.6, and C.6 had almost the same PC1 and PC2 value in the scatter plot. During the incubation of batches A.6, B.6, and C.6, no changes were observed in the PC1 and PC2, indicating only a small change in the microbial diversity.
Principal component analysis scatter plot of DGGE profiles (Fig. 6) of sixth-generation transfers A.6 cultured with 11 μM tNP till t = 15 days and thereafter with 1 mM tNP, B.6 and C.6 incubated with 11 μM tNP, and fourth-generation transfer D.4 incubated with 1 mM tNP at different time intervals
The DGGE pattern of the sample of batch D.4, which is a fourth-generation transfer cultured with 1 mM tNP (above saturation), contained 14 bands. The Shannon–Weaver index for microbial diversity of sample of D.4 was 1.05. This indicates that the microbial diversity in sample D.4 was less than in the samples of batches A.6 to C.6. The pattern showed similarities with the patterns of the samples of batches A.6 to C.6, but there were also some different dominating bands present. The PCA results of the DGGE profiles (Fig. 6) show that the data point of sample D.4 did not cluster with the results of batches A.6, B.6, and C.6. This indicates that incubation with a tNP concentration above maximum solubility gives a different microbial population compared to incubation with 11 μM tNP concentration.
Microbial population composition
As the microbial composition of A.6 did not change during the incubation time and was similar to batches B.6 and C.6, a clone library was made of batch A.6. The DNA extract from t = 15 days was used when nitro-nonylphenol was formed. A second clone library was made from batch D.4 at t = 55 days, which showed a different microbial composition on the DGGE pattern. Batch D.4 was cultured with 1 mM NP, whereas A.6, B.6, and C.6 were cultured with 15 μM tNP. Respectively, 70 and 49 clones were obtained from batches A.6 and D.4 (Table 1). The most dominant clones of A.6 were related to Nitrosomonas genus (36 out of 70 clones), which are ammonium-oxidizing bacteria (Stein et al. 2007). Of these 36 clones, 18 were closely related to Nitrosomonas euthropha (GenBank accession number AY123795) with a maximum 16S rRNA gene identity of 91–100%. Another 16 clones were closely related to N. euthropha C91 with a maximum identity of 99%. Alignment of the sequences of N. euthropha (GenBank accession number AY123795) and N. euthropha C91 showed a 99% similarity between these two strains. In the clone library of batch D.4, clones were also identified that are related to N. euthropha (two out of 49) and N. euthropha C91 (four out of 49); however, they were less abundantly present than in the clone library of batch A.6. Three clones were related to Nitrosomonas sp. GC22 with a similarity of 96–99%.
Clones related to the sequence of Beta proteobacterium C14 JRPA-2007 (eight out of 70 clones) were also dominantly present in the clone library of batch A6. B. proteobacterium C14 JRPA-2007 sequence belongs to the Alcaligenacea family and is closely related to Alcaligenes sp. Ic4, a 3-chlorobenzoate degrading microorganism (Krooneman et al. 1996). Clones (four out of 49 clones) related to B. proteobacterium C14 JRPA-2007 were also present in the clone library of batch D.4.
The most dominating clones in the clone library from batch D.4 belonged to the family Comamonadacea (28 out of 49 clones). Thirteen clones were closely related to Acidovorax sp. PD-10 (92–99% similarity) and 11 clones to Alicycliphillus sp. R-24604 (96–99% similarity). Acidovorax sp. PD-10 is closely related to the species Acidovorax avenae isolate C1 (AF508114), which is a phenol-degrading, nitrate-reducing bacterium (Baek et al. 2003). Alignment of the sequences of Alicycliphillus sp. R-24604 (AM084015) and A. avenae isolate C1 (AB195159) gave 99% similarity of these two strains. This indicates that Alicycliphillus sp. R-24604 is very closely related to A. avenae isolate C1 as well. Clones related to Acidovorax sp. PD-10 and Alicycliphillus sp. R-24604 were also identified in the clone library of batch A.6 and were less dominantly present than in the clone library of batch D.4.
Discussion
The mixture of branched NP isomers that were present in the polluted sediment can be biodegraded under aerobic conditions by the autochthon microorganisms. The biodegradation of NP in our sediment was much faster compared to the aerobic degradation of NP in sediment from the Erren river in China (Yuan et al. 2004). Within 8 days, approximately 4.5 μM NP was biodegraded, whereas biodegradation of 9 nM in Erren river sediment took 84 days. Adaptation of the Erren sediment to NP degradation increased the biodegradation to 28 days. Faster biodegradation in our sediment is due to adaptation of the bacteria to NP because our used sediment was already polluted with NP.
Subsequent transfers of the microbial population from our sediment enlarged the lag phase compared to the first generation and increased the half-life time of tNP. Often, transfers of a culture result in the enrichment of the involved microorganisms in the degradation process and a decrease of the half-life time. Due to our transfers, the originally present sediment with some river water was diluted out, and the fifth-generation batches hardly contained any sediment. This diluting out of the sediment might play a role for the bacteria, either as carrier material of for essential nutrients.
The biodegradation by microorganisms in the sediment of NP, both branched and linear NP, occurs via the formation of a nitro-nonylphenol metabolite. The attached NO2-group in the formed nitro-nonylphenol originates from ammonium and is a biotic process in our experiments. The formation of nitro-nonylphenol, with the nitro-group attached to the phenolic ring, was also observed in biodegradation studies with agricultural soils (Zhang et al. 2009) and a mixture of agricultural soils with sewage sludge (Telscher et al. 2005; Zhang et al. 2009). This indicates that the formation of nitro-nonylphenol is relevant in the biodegradation of tNP. The formed nitro-nonylphenol in our experiments was further degraded, which was also observed in agricultural soils (Zhang et al. 2009). In these soils, the formed nitro-nonylphenol was not completely degraded; 13% of the initial applied NP remained in the sediment as a nitro-metabolite. In our experiments, the nitro-nonylphenol was only completely degraded when 11 μM tNP was applied. Higher concentrations (1 mM) led to accumulation of the nitro-metabolites. Despite this incomplete degradation, biodegradation of NP in this sediment leads to a reduction of the estrogenic effect of the sediment (de Weert et al. 2008), as formed nitro-nonylphenol metabolites are less toxic (85% less) than NP (Kammann et al. 2009); (Zhang et al. 2009)
Molecular analyses showed the presence of ammonium-oxidizing species Nitrosomonas eutropha and N. eutropha C91. They can be responsible for oxidation of the ammonium to NO −2 , which is attached to the phenolic ring of NP. The sequences of N. eutropha C91 are closely related to the sequence of Nitrosomonas europaea (Koops et al. 1991). N. europaea is a strain that oxidizes nonsubstituted alkanes (Hyman et al. 1988) and is able to co-oxidize aromatic compounds in the presence of ammonium (Keener and Arp 1994). In our batches, the formed nitro-nonylphenol was further degraded. As N. europaea has not been described to catalyze ring fission of aromatic compounds, this indicates that these organisms are not involved in the degradation of the aromatic ring in our experiments. If Nitrosomonas species form the nitro-nonylphenol but are not able to cleave the aromatic ring structure, other species must be involved in further biodegradation of the nitro-nonylphenol. Both clone libraries of the batches cultured with 11 μM or 1 mM NP contain sequences of species related to B. proteobacterium C14 JRPA-2007 (95–99% similarity). B. proteobacterium C14 JRPA-2007 belongs to the Alcaligenes genus. The Alcaligenes genus contains species able to degrade ortho-nitro-phenol (Xiao et al. 2007). Other strains, which are related to nitro-phenol degradation that is a comparable structure as the nitro-phenol part in our in nitro-nonylphenol metabolites, are not identified in both clone libraries.
Both clone libraries contain sequences related to nitrate-reducing Acidovorax sp. PD-10 and Alicycliphillus sp. R24604, which are abundantly present. Both species are closely related to A. avenae isolate C1, a nitrate-reducing, phenol-degrading bacterium (Baek et al. 2003). As our batches are aerobic, it is not likely that the nitrate-reducing strains related to Acidovorax sp. PD-10 and Alicycliphillus sp. R24604 are involved in the formation of nitro-nonylphenol. Furthermore, the NO2-group in the nitro-nonylphenol originates from ammonium instead of nitrate. Biodegradation of phenol by A. avenae isolate C1 is suggested to occur via cleavage of the ring structure (Baek et al. 2003), and this ring cleavage might also occur for NP, which is also proposed for the NP degradation by Pseudomonas species (Soares et al. 2003). The abundant presence of species related to the phenol-degrading A. avenae isolate C1 after several transfers and the presence of NP as only carbon source suggest that the biodegradation of NP occurs via the phenolic degradation pathway. Unfortunately, we have no direct evidence for this pathway, and this should be further investigated.
When tNP is degraded by Sphingomonas and Sphingobium species via ipso-hydroxylation (Gabriel et al. 2005a; Kohler et al. 2008), a shift in isomeric composition of the tNP occurs. We did not observe such a shift in the isomeric composition during the biodegradation of the mixture of branched NP isomers in the first-generation batches. Furthermore, sequences related to Sphingomonas and Sphingobium species were not identified in our batches. Both the absence of the shift in isomeric composition and the absence of strains involved in the NP degradation via ipso-hydroxylation gives a strong indication that biodegradation of NP via ipso-hydroxylation in our batches is not relevant. The microorganisms identified in this study are not related to known NP-degrading bacteria such as Sphingomonas, Sphingobium, Stenotrophomonas, or Pseudomonas species (Tanghe et al. 1999; Soares et al. 2003; Ushiba et al. 2003; Gabriel et al. 2005a). This indicates that aerobic NP degradation can be performed by a wide range of microbial species and via different degradation pathways. Transfers of the bacterial population will give a different NP-degrading population as the original population in the sediment because some involved bacteria will get lost during enrichment. The untouched environment might, therefore, contain even more bacteria which are involved in the biodegradation of NP than we have identified in our cultures. The involvement of a wide range of microorganisms related to NP biodegradation enlarges the possible risk reduction of NP in the environment by biodegradation under aerobic conditions.
References
Baek S-H, Kim K-H, Yin C-R, Jeon CO, Im W-T, Kim K-K, Lee S-T (2003) Isolation and characterization of bacteria capable of degrading phenol and reducing nitrate under low-oxygen conditions. Curr Microbiol 47:462–466
Chang BV, Yu CH, Yuan SY (2004) Degradation of nonylphenol by anaerobic microorganisms from river sediment. Chemosphere 55:493–500
Corvini PFX, Schäffer A, Schlosser D (2006a) Microbial degradation of nonylphenol and other alkylphenols—our evolving review. Appl Microbiol Biotechnol 72:223–243
Corvini PFX, Hollender J, Schumacher S, Prell J, Hommes G, Priefer U, Vinken R, Schaffer A (2006b) The degradation of alpha-quaternary nonylphenol isomers by Sphingomonas sp. strain TTNP3 involves a type II ipso-substitution mechanism. Appl Microbiol Biotechnol 70:114–122
Corvini PFX, Meesters RJW, Mundt M, Schäffer A, Schmidt B, Schröder HF, Verstraete W, Vinken R, Hollender J (2007) Contribution to the detection and identification of oxidation metabolites of nonylphenol in Sphingomonas sp. strain TTNP3. Biodegradation 18:233–245
Hesselsoe M, Jensen D, Skals K, Olesen T, Moldrup P, Roslev P, Mortensen GK, Hendriksen K (2001) Degradation of 4-nonylphenol in homogeneous and nonhomogeneous mixtures of soil and sewage sludge. Environ Sci Technol 35:3695–3700
de Vries YP, Takahara Y, Ikunaga Y, Ushiba Y, Hasegawa M, Kasahara Y, Shimomura H, Hayashi S, Hirai Y, Ohta H (2001) Organic nutrient-dependent degradation of branched nonylphenol by Sphingomonas sp. YT isolated from river sediment samples. Microbes Environ 16:240–249
de Weert J, De la Cal A, Hvd B, Murk AJ, Langenhoff AAM, Rijnaarts HHM, Grotenhuis JTC (2008) Bioavailability and biodegradation of nonylphenol in sediment determined with chemical and bioanalysis. Environ Toxicol Chem 27:778–785
Ekelund R, Granmo A, Magnusson K, Berggren M (1993) Biodegradation of 4-nonylphenol in seawater and sediment. Environ Pollut 79:59–61
Gabriel FLP, Giger W, Guenther K, Kohler H-PE (2005a) Differential degradation of nonylphenol isomers by Sphingomonas xenophaga Bayram. Appl Environ Microbiol 71:1123–1129
Gabriel FLP, Heidlberger A, Rentsch D, Giger W, Guenther K, Kohler H-PE (2005b) A novel metabolic pathway for degradation of 4-nonylphenol environmental contaminants by Sphingomonas xenophaga Bayram. J Biol Chem 22:15526–15533
Giger W, Brunner PH, Schaffner C (1984) 4-Nonylphenol in sewage sludge: accumulation of toxic metabolites from nonionic surfactants. Science 225:623–625
Heemken OP, Reincke H, Stachel B, Theobald N (2001) The occurrence of xenoestrogens in the Elbe River and the North Sea. Chemosphere 45:245–259
Hou S-G, Sun H-W, Goao Y (2006) Sorption of small metabolites of nonylphenol polyethoxylates in single and complex systems on aquatic suspended particulate matter. Chemosphere 63:31–38
Hyman MR, Murton IB, Arp DJ (1988) Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl Environ Microbiol 54:3187–3190
Kammann U, Vobach M, Wosniok W, Schäffer A, Telscher MJH (2009) Acute toxicity of 353-nonylphenol and its metabolites for zebrafish embryos. Environ Sci Pollut Res 16:227–231
Keener WK, Arp DJ (1994) Transformation of aromatic compounds by Nitrosomonas europaea. Appl Environ Microbiol 60:1914–1920
Kohler H-PE, Gabriel FLP, Giger W (2008) Ipso-substitution—a novel pathway for microbial metabolism of endocrine-disrupting 4-nonylphenols, 4-alkoxyphenols, and bisphenol A. Chimia 62:358–363
Koops H-P, Böttcher B, Möller U, Pommerening-Röser A (1991) Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. J Gen Microbiol 13:1689–1699
Krooneman J, Wieringa EBA, Moore ERB, Gerritse J, Prins RA, Gottschal JC (1996) Isolation of Alicaligenes sp. strain L6 at low oxygen concentrations and degradation of 3-chlorobenzoate via a pathway not involving (chloro)catechols. Appl Environ Microbiol 62:2427–2434
Lacorte S, Raldúa D, Martínez E, Navarro A, Diez S, Bayona JM, Barceló D (2006) Pilot survey of a broad range of priority pollutants in sediment and fish from the Ebro River basin (NE Spain). Environ Pollut 140:471–482
LeBlanc GA, Rider CV (2000) Embryotoxicity of the alkylphenol degradation product 4-nonylphenol to the crustacean Daphnia magna. Environ Health Perspect 108:1133–1138
Navarro A, Endo S, Gocht T, Barth JAC, Lacorte S, Barcelo D, Grathwohl P (2009) Sorption of alkylphenols on Ebro River sediments: comparing isotherms with field observations in river water and sediments. Environ Pollut 157:698–703
Petrovic M, Solé M, López de Alda MJ, Barcelo D (2002) Endocrine disruptors in sewage treatment plants, receiving river waters, and sediments: integration of chemical analysis and biological effects on feral carp. Environ Toxicol Chem 212:2146–2156
Servos MR (1999) Review of the aquatic toxicity, estrogenic responses and bioaccumulation of alkylphenols and alkylphenol polyethoxylates. Water Qual Res J Can 34:123–177
Shannon C, Weaver W (1949) The mathematical theory information. University of Illinois Press, Urbana
Snyder AS, Keith TL, Pierens SL, Snyder EM, Giesy JP (2001) Bioconcentration of nonylphenol in fathead minnows (Pimphales promelas). Chemosphere 44:1697–1702
Soares A, Guieysse B, Delgado A, Mattiasson B (2003) Aerobic biodegradation of nonylphenol by cold adapted bacteria. Biotechnol Lett 25:731–738
Stein LY, Arp DJ, Berube PM, Chain SG, Hauser L, Jetten MSM, Klotz MG, Larimer FW, Norton JM, HJMod C, Shin M, Wei X (2007) Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ Microbiol 9:2993–3007
Stephanou E, Giger W (1982) Persistent organic chemicals in sewage effluents. 2. Quantitative determination of nonylphenols and nonylphenolethoxylates by glass capillary gas chromatography. Environ Sci Technol 16:800–805
Tanghe T, Dhooge W, Verstaete W (1999) Isolation of a bacterial strain able to degrade branched nonylphenol. Appl Environ Microbiol 65:746–751
Telscher MJH, Schuller U, Schmidt B, Schaffer A (2005) Occurrence of nitro metabolite of a defined nonylphenol isomer in soil/sewage sludge mixtures. Environ Sci Technol 39:7896–7900
Thiele B, Heinke V, Kleist E, Guenther K (2004) Contribution of the structural elucidation of 10 isomers of technical p-nonylphenol. Environ Sci Technol 38:3405–3411
Tros ME, Schraa G, Zehnder AJB (1996) Transformation of low concentrations of 3-chlorobenzoate by Pseudomonas sp. strain B13; kinetics and residual concentrations. Appl Environ Microbiol 62:437–442
Tyler CR, Jobling S, Sumpter JP (1998) Endocrine disruption in wildlife: a critical review of the evidence. Crit Rev Toxicol 28:319–361
Ushiba Y, Takahara Y, Ohta H (2003) Sphingomonas amiense sp. nov., a novel nonylphenol-degrading bacterium isolated from a river sediment. Int J Syst Evol Microbiol 53:2045–2048
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16s ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wheeler TF, Hein JR, Latorre MR, James B (1997) Mass spectral characterization of p-nonylphenol isomers using high-resolution capillary GC-MS. J Chromatogr Sci 35:19–30
Xiao Y, Zhang J-J, Liu H, Zhou N-Y (2007) Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. Strain NyZ215. J Bacteriol 189:6587–6593
Yang F-X, Xu Y, Wen S (2005) Endocrine-disrupting effects of nonylphenol, bisphenol A, and p, p-DDE on Rana nigromaculata tadpoles. Bull Environ Contam Toxicol 75:1168–1175
Yu Z, Morrison M (2004) Comparisons of different hypervariable regions of rrS genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 70:4800–4806
Yuan SY, Yu CH, Chang BV (2004) Biodegradation of nonylphenol in river sediment. Environ Pollut 127:425–430
Zhang H, Spiteller M, Guenther K, Boehmler G, Zuehlke S (2009) Degradation of a chiral nonylphenol isomer in two agricultural soils. Environ Pollut 157:1904–1910
Acknowledgment
This research was financially supported by the EU FP6 Aquaterra (project number 505428 GOCE). The authors want to thank Maarten Posthumus of the Department of Organic Chemistry of Wageningen University, The Netherlands for his help and the accurate mass weight analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
De Weert, J., Viñas, M., Grotenhuis, T. et al. Aerobic nonylphenol degradation and nitro-nonylphenol formation by microbial cultures from sediments. Appl Microbiol Biotechnol 86, 761–771 (2010). https://doi.org/10.1007/s00253-009-2394-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2394-9