Abstract
With the completion of several genome sequences for parasitic protozoa, research in molecular parasitology entered the “post-genomic” era. Accompanied by global transcriptome and proteome analysis, huge datasets have been generated that have added many novel candidates to the list of drug and vaccine targets. The challenge is now to validate these factors and to bring science back to the bench to perform a detailed characterization. In some parasites, like Trypanosoma brucei, high-throughput genetic screens have been established using RNA interference [for a detailed review, see Motyka and Englund (2004)]. In most protozoan parasites, however, more time-consuming approaches have to be employed to identify and characterize the function of promising candidates in detail. This review aims to summarize the status of molecular genetic tools available for a variety of protozoan pathogens and discuss how they can be implemented to advance our understanding of parasite biology.
Similar content being viewed by others
Introduction
Diseases caused by parasitic protozoa pose a significant burden on public and veterinary health worldwide, mainly affecting developing countries and deprived communities. Only a few drugs for diseases like malaria, sleeping sickness, leishmaniasis, and Chagas disease are available and, typically, suffer shortcomings of toxicity and ineffectiveness due to the development of resistance. Therefore, novel drug and vaccine candidates are urgently needed that are safer, more effective, and affordable. To fulfill this task, it is important to unravel the complex and fascinating biology of this organisms that led to many surprising and novel findings like RNA editing in kinetoplastida (Simpson et al. 2003) or the apicoplast, a newly identified organelle in apicomplexan parasites that is a promising drug target (Waller and McFadden 2005).
Ongoing efforts to complete microbial genome sequencing projects, along with transcriptome and proteome characterization, have generated an impressive amount of data waiting to be analyzed in more detail. Consequently, a significant challenge in the field now is to decide which of the identified candidate proteins are indeed promising targets for vaccine and drug development.
The general aim in the post-genomic era is to convert our knowledge of the genome, transcriptome, and proteome into knowledge of the “phenome” (the physical totality of all traits of an organism; Mahner and Kary 1997). One possible way to reach this goal is to generate a list of promising candidates and characterize them one by one using reverse genetic tools. However, many hypothetical proteins with unknown function that could be of great significance might be omitted from this analysis. Therefore, forward genetic screens for certain phenotypes will help identify and characterize novel factors involved in different processes pertinent to the life cycles of the various parasitic pathogens. This review aims to give an overview of the different approaches currently employed to perform gene/function analysis in select protozoan parasites. It is important to keep in mind that “protozoan parasites” do not necessarily define a group of related organisms. In fact, these parasites represent many highly diverged phyla and, thus, have evolved numerous different survival strategies. This typically implies that new parasite tools must be developed, tailored to fit the unique biology and genomic constitution of each pathogenic group. In this paper, we will focus on molecular tools employed in some representative model organisms, including Giardia lamblia, Entamoeba histolytica, Trypanosoma brucei, Toxoplasma gondii, and Plasmodium spec. An overview of established tools discussed in this review is given in Table 1.
Parasite genomes: an explosion of data
Research into parasite biology has changed dramatically with the publication of numerous genomes including the three main pathogenic species of trypanosomatida (Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major (Gardner et al. 2002; El-Sayed et al. 2005a, b; Ivens et al. 2005), the archamoeba E. histolytica (Loftus et al. 2005), the apicomplexan parasites P. falciparum, P. yoelii yoelii, and other Plasmodium species (Carlton et al. 2002), Cryptosporidium parvum (Abrahamsen et al. 2004), C. hominis (Xu et al. 2004), Theileria parva (Gardner et al. 2005), T. annulata (Pain et al. 2005), and Toxoplasma gondii (http://www.ToxoDB.org). In addition, the genome sequencing effort of the amitochondriate G. lamblia is nearing completion. These immense datasets not only allow the identification of virtually any parasite gene but also permit comparative genomic analysis. Comparative genomics is providing substantial insight into the evolution of parasites and species-specific phenomena related to the unique requirements of a particular microbe (El-Sayed et al. 2005a, b; Kooij et al. 2005).
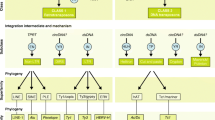
The combined efforts of multiple groups have provided the community with invaluable insight into the basic composition and biology of diverse parasites. Until recently, research has focused on a relatively limited number of candidate factors that have been identified before the genomic era. Although characterised in great detail, these candidates often failed to live up to expectations. Now, the community is faced with a huge (and still expanding) list of new predicted gene products, and the daunting task of deciding which one(s) warrant priority analysis. While the combination of data available from comparative genomics, transcriptomics, and proteomics provides clues to the putative functions of some predicted gene products, the validation of these as pharmacological or vaccine targets mandates the establishment of high-throughput assays; these goals may be reached through a combination of reverse and forward genetics tools (Fig. 1).
Transfection of protozoan parasites opens the way to molecular biology
A key breakthrough facilitating the use of any given organism as a molecular model system is the development of efficient transfection techniques. Whereas successful transfection of yeast was described in 1978 using chemical transformation protocols (Hinnen et al. 1978), effective transfection of protozoan parasites was not achieved until nearly a decade later, the first being described for Trypanosoma brucei in 1987 (Eid and Sollner-Webb 1987). Shortly after this milestone, similar transfection systems have been established for other kinetoplastida (Clayton 1999), E. histolytica (Nickel and Tannich 1994; Purdy et al. 1994), G. lamblia (Yee and Nash 1995), and apicomplexan parasites, including many Plasmodium species (Soldati and Boothroyd 1993; van Dijk et al. 1995; Wu et al. 1995).
With the establishment of diverse selectable markers, protozoan parasites became accessible to genetic manipulation, thereby allowing the generation of transgenic parasites. In addition, the successful expression of fluorescent markers has allowed a detailed look at protein trafficking mechanisms, dynamic protein localisation (Gubbels and Striepen 2004), and in vivo imaging of parasites (Amino et al. 2005).
Transient and stable transfection technology clearly opened the way for an in-depth analysis of fundamental parasite biology. However, additional tools had to be developed to allow the characterization of essential genes. Disruption of nonessential genes is relatively straightforward in organisms with culturable haploid stages during which homologous recombination can take place, e.g., asexual stages of Toxoplasma gondii (Kim et al. 1993), P. falciparum (Crabb et al. 1997), or P. berghei (van Dijk et al. 1995; Wu et al. 1996). This approach is quite time-consuming in parasites with a diploid genome, like trypanosomatids (Hariharan et al. 1993), and nearly impossible in G. lamblia or E. histolytica, which have two diploid nuclei. In this cases, overexpression of dominant negative proteins can lead to a phenotype of a null mutant as, for example, in the case of overexpression of light meromyosin that led to a myosin II null phenotype (Arhets et al. 1998).
Among the most interesting factors in the genome of parasites are genes with essential functions that are expressed during the entire life cycle of the parasite; however, such genes cannot be disrupted easily using conventional approaches. Alternative strategies that permitted controlled or conditional reduction of the expression level of the gene of interest had to be devised. A number of independent groups have succeeded in establishing the techniques aimed at controlled genomic rearrangements, transcriptional regulation, or post-transcriptional regulation.
Genome manipulation using site-specific recombination
One of the attractive possibilities to generate mutants for an essential gene is to employ site-specific recombination systems, such as the Cre/lox, Flp/FRT, or related systems. The basic principle of these systems is to flank a gene of interest by specific recognition elements for the respective recombinase. After activation of the recombinase, the target locus is modified, resulting either in the excision or inversion of the respective region (Fig. 2a).
Different strategies to manipulate the genome and gene expression in protozoan parasites. a A site-specific recombination system allows removal or inversion of a target DNA sequence flanked by recognition sites for the recombinase (Flp or Cre). b Employing regulatory elements derived from the Tet operon (tetO) of E. coli, two different systems have been successfully established in protozoan parasites. The repression system (upper panel) works mainly by interference with initiation of transcription by placing upstream TetO sequences proximal to the transcription start. In the absence of the inducer (tetracycline derivatives), TetR binds tightly to TetO and blocks transcription. In the transactivator system (lower panel), a transactivation domain is fused to TetR so that it can interact with the basic transcription machinery. Upon binding to TetO sequences placed upstream of a silent minimal promoter, this chimeric protein is capable of recruiting the transcription machinery to the promoter, thereby activating transcription. In the presence of the inducer, the TetR–TA fusion is not binding to TetO, thereby turning the promoter off. c Two different strategies employing the Tet transactivator system are currently being used for the generation of conditional mutants in apicomplexan parasites (see text for detailed explanation)
The difficulty of these strategies lies in triggering, either temporally or locally, the recombination event. In mice, for example, this can be achieved by controlling the expression of the recombinase using tissue-specific or inducible promoters that results in a location or time-dependent disruption of the locus [for a recent review, see Branda and Dymecki (2004)]. Alternatively, the usage of ligand-controlled recombinases has been reported to add temporal control in such systems (Metzger et al. 1995).
Different groups have attempted to adapt this strategy for use in protozoan parasites. Among the first reports are applications of the Cre-lox system for Toxoplasma gondii (Brecht et al. 1999) and trypanosomatids (Barrett et al. 2004). In both cases, however, the problem of tight regulation of Cre expression appears to compromise the fidelity of the system. In Toxoplasma gondii, for example, the expression of Cre under a stage-specific (and silent) promoter still resulted in recombination events, indicating that even basal expression of Cre is sufficient to produce recombination (Brecht et al. 1999). We have recently tried to establish a tetracycline-inducible Cre-lox system, but even this did not appear to circumvent the problems associated with “leaky” Cre expression under supposedly silenced conditions (Kessler and Meissner, unpublished results).
A very promising approach employing the Flp/FRT-system in P. berghei (Carvalho et al. 2004) has been described. In this case, the regulation of the recombination event was achieved by creating two independent parasite strains: one with the locus of interest flanked with the recombinase recognition sites (FRT) and one expressing the Flp-recombinase itself. Upon cross-fertilization of each strain in the mosquito (where the sexual cycle takes place), offspring are generated according to Mendelian laws, and the parasites with the deleted locus can be selected. This approach might have some limitations considering the range of applications at its current stage. Although candidate genes can be efficiently removed upon cross-fertilization, the resulting phenotype needs to be analyzed in the respective stage where the gene normally functions. For example, if the candidate gene is essential in asexual blood stages, no viable parasites will be easily isolated, thus, hampering a detailed phenotypic characterization.
Recently, a novel approach for stable transfection of P. falciparum has been established using the integrase Bxb1 of mycobacteriophage, which allows efficient and rapid site-specific integration of a plasmid containing the recognition site attB into a locus containing the attP site (which needs to be engineered using standard technologies; Nkrumah et al. 2006). Indeed, this approach will ease the task of generating transgenic parasite strains in the near future.
Regulation of parasite gene expression
A greater understanding of how protozoan parasites regulate gene expression will facilitate the development of transgenic lines aimed to interrogate the function of genes of interest. Most eukaryotes regulate gene expression primarily at the transcriptional level. However, comparative genomic analysis of protozoan parasites reveals an unexpected paucity of conventional transcription factors (Meissner and Soldati 2005). In contrast, virtually all epigenetic and chromatin-remodeling machineries are conserved in these parasites (Sullivan et al. 2006). These observations suggest that transcriptional regulation may contain some fundamental differences in these early-branching eukaryotic cells. An extreme example of this is exemplified by the kinetoplastida. As in all eukaryotes, these parasites have three different RNA polymerases. However, whereas in all other eukaryotes, RNA polymerase I transcribes, almost exclusively, ribosomal DNA and polymerase II protein-coding genes; this clear functional distinction has been overcome in kinetoplastida. Interestingly, most genes are transcribed as polycistronic pre-messenger RNA (mRNA) and are subsequently processed by trans-splicing (which adds a capped 5′end to the mRNA) and poly-adenylation. This uncoupling of 5′ capping and transcription of protein-coding genes allows efficient transcription by RNA polymerase I. Due to polycistronic transcription, the main level of regulation of gene expression has shifted to post-transcriptional regulation via control of RNA stability [for a review, see Clayton (2002) and Teixeira and daRocha (2003)].
Despite the apparent lack of transcription factors in protozoan parasites in general, tight transcriptional regulation has been demonstrated for many different genes in apicomplexans, E. histolytica and G. lamblia (Vanacova et al. 2003; Mirelman et al. 2006). The analysis of apicomplexan transcriptomes by serial analysis of gene expression (SAGE) further support that transcriptional mechanisms play a key role in governing parasite development (Patankar et al. 2001; Radke et al. 2005). The identification and characterization of cis-acting elements also argue that these parasites have evolved a complex gene regulation network, further supported by whole genome transcriptome analysis for P. falciparum (Bozdech et al. 2003; Le Roch et al. 2003) and by analysis of transcriptional changes during Toxoplasma gondii differentiation using microarrays (Cleary et al. 2002). Elucidating the factors that are likely to be binding to these cis-acting elements in apicomplexan promoters is an area of intense investigation.
As alluded to above, epigenetic modes of transcription regulation also appear to make substantial contributions to parasite gene expression. Histone modifications have been demonstrated to play an important role in the transcriptional regulation involved in antigenic variation in P. falciparum, Trypanosoma brucei, and G. lamblia [for a recent review, see Sullivan et al. (2006)]. Histone modifications have also been correlated with stage-specific transcription during differentiation in Toxoplasma gondii (Saksouk et al. 2005).
It has been speculated that the lack of transcriptional regulatory proteins dictate that the few present are used in a more combinatorial manner to achieve gene regulation (van Noort and Huynen 2006). Alternatively, it is plausible that protozoan parasites employ noncoding RNAs (ncRNAs) for both transcriptional and post-transcriptional regulation. An increasing number of functions have been assigned to ncRNAs in other organisms including direct regulation of transcription in trans by targeting transcriptional activators and repressors, general transcription factors, and RNA polymerase II [for a recent review, see Goodrich and Kugel (2006)]. Given the large amount of ncRNAs and antisense transcripts identified so far in protozoan parasites, e.g., Plasmodium (Gunasekera et al. 2004) and G. lamblia (Elmendorf et al. 2001), it is possible that similar mechanisms of transcriptional regulation may exist. Recently, it has been demonstrated in E. histolytica that transcriptional gene silencing can be induced artificially by the expression of a small heterochromatic RNA containing the 5′ end of a short interspersed nuclear element (SINE1; Bracha et al. 2006). It will be interesting to assess if similar mechanisms of transcriptional silencing also occur in other protozoan parasites.
While gene expression most certainly is controlled at the transcriptional level, there are important lines of evidence showing that translational regulation may also come into play. For example, translational control mediates the developmental regulation of the Trypanosoma brucei Nrk protein kinase (Gale et al. 1994). Puf proteins, which regulate translation and RNA stability, have been documented in P. falciparum (Cui et al. 2002). A translational repressor complex involved in the regulation of gametocyte-specific expression in P. berghei has also been described (Mair et al. 2006). In Toxoplasma gondii, eIF2α becomes phosphorylated in response to stress, which is another well-known event that regulates translation in other species (Sullivan et al. 2004).
Inducible transcriptional regulation
Although little is known about the detailed mechanisms of transcriptional regulation, numerous groups have succeeded in establishing inducible transcription systems in protozoan parasites. In general, an inducible system needs to be efficient and highly specific for the gene of interest and should not cause any pleiotropic effects on endogenous genes. The usage of regulatory elements from unrelated organisms, such as bacteria, has proved to be a promising strategy in other eukaryotes. The first attempts to modulate transcription in mammalian cell lines made use of a prokaryotic repressor/operator system based on the lac operon of Escherichia coli (Deuschle et al. 1986; Brown et al. 1987; Hu and Davidson 1987). The integration of lac operator sequences within the context of a strong constitutive promoter leads to IPTG-dependent control of transcription initiation in a line expressing the lac repressor (lacI). An analogous system making use of the tetracycline (tet) operon of E. coli has been established in various organisms, including plants (Gatz and Quail 1988; Gatz et al. 1992) and protozoan parasites. Shortly after the first report describing tight control of gene expression using the Tet-repressor/operator system in Trypanosoma brucei (Wirtz and Clayton 1995) appeared, various groups established similar systems in other protozoan parasites, including E. histolytica (Hamann et al. 1997; Ramakrishnan et al. 1997), G. lamblia (Sun and Tai 2000), L. donovani (Yan et al. 2001), and Toxoplasma gondii (Meissner et al. 2001). The system established in Trypanosoma brucei proved very powerful, especially with some refinements such as the inclusion of T7 RNA polymerases for transcription, which can be blocked efficiently by TetR (Wirtz et al. 1998), and the combination with RNA interference that allows inducible down-regulation of protein expression (Wang et al. 2000). Similarly, in G. lamblia and E. histolytica, effective reduction of protein expression has been achieved by inducible expression of antisense transcripts (Sahoo et al. 2003; Touz et al. 2005; Vats et al. 2005). To date, the feasibility of using the repressor system in Toxoplasma gondii for similar approaches remains to be tested. Our results indicate that the current system displays a relatively high tendency of losing efficient gene regulation, especially when toxic or essential genes are put under control of this system (our own unpublished results). Recently, a slightly modified version of the repressor system employing a YFP-TetR fusion has been reported for Toxoplasma gondii (van Poppel et al. 2006), but it remains to be shown if this modification results in a more robust inducible system without reversion effects.
In higher eukaryotes, a transactivator-based system is used more broadly than repressor-based systems. In this case, a fusion between a TetR and the viral protein 16 (VP16) of the Herpes simplex virus converts the former into an efficient tetracycline-dependent transactivator (tTA), thereby allowing strong activation of an otherwise silent (minimal) promoter placed downstream of tandemly repeated TetO sequences (Gossen and Bujard 1992). This system has since been optimized in many different ways and is used commonly in cultured cells as well as in transgenic mice [for a review, see Corbel and Rossi (2002)]. The two essential components of this system (the transactivator and the inducible minimal promoter) are broadly functional in eukaryotes. However, early attempts to employ the original tTA system in Toxoplasma gondii failed because the transactivation domain (TD) of VP16 and the minimal promoter derived from cytomegalovirus (pCMV) both appeared to be non-functional when introduced in the parasite (Meissner et al. 2001). Hence, components functional in apicomplexan parasites needed to be identified to generate a working Tet-transactivator system in apicomplexan parasites. Previous characterization of the promoter region for the major surface antigen SAG1 in Toxoplasma gondii led to the identification of a heptamer repeat acting in cis. Upon deletion of this element, the minimal SAG1 promoter is rendered virtually silent (Soldati and Boothroyd 1995). Therefore, it was reasoned that substitution of the heptamer repeats of pSAG1 with TetO sequences should result in a Tet-responsive promoter that can be activated upon binding of a TetR–TD fusion. To isolate a TD functional in Toxoplasma gondii, a screen based on random insertional mutagenesis was performed, leading to the identification of two artificial TDs (termed TATi-1 and TATi-2) that allowed efficient regulation of transcription in the parasite (Meissner et al. 2002). Subsequently, it was shown that the identified TDs are also capable of activating transcription in P. falciparum (Meissner et al. 2005).
While the transactivator system established in P. falciparum might need further optimization for the characterization of essential genes, this system has been successfully employed numerous times to facilitate the characterization of essential genes in Toxoplasma gondii (Meissner et al. 2002; Mital et al. 2005; Huynh and Carruthers 2006; Mazumdar et al. 2006). Two strategies for the establishment of conditional mutants are currently employed (Fig. 2). The first approach requires the stable transfection of a second, inducible copy of the gene of interest into the TATi-expressing strain. After selection of a clonal parasite strain harboring expression levels of the transgenic copy matching that of the endogenous gene (assessed by RT-PCR or immunoblot), the endogenous copy of the target gene is disrupted by homologous recombination. This results in a conditional, inducible mutant for this gene. While this approach is more time consuming, it has a higher probability of success.
The second, more straightforward approach has been successfully used in our laboratory for low abundance genes (Agop-Nersesian and Meissner, unpublished results). The employment of a vector that allows direct exchange of the endogenous promoter with the Tet-inducible promoter via homologous recombination directly puts the endogenous gene under control of the Tet system. The advantage of this approach is that the conditional mutant can be established in a single transfection within a few weeks (versus months, in case of the first strategy). However, this strategy might be problematic in the case of strongly expressed genes that may be essential. In this event, the direct exchange of a strong endogenous promoter for the relatively weak Tet-inducible promoter might not be possible.
Although increasing the number of TetO sequences placed upstream of the minimal promoter does not significantly increase expression levels (our own unpublished results), the usage of different minimal promoters might facilitate a higher range of regulation obtained. Alternatively, optimization of the current TD or identification of novel, more efficient TDs may lead to a more efficient regulatory system. A modified screen aimed at the identification of novel TDs has yielded several independent candidates showing the activation of the inducible promoter (Klaus and Meissner, unpublished results). The identification of further TDs is not only worthwhile in terms of tool development, but will also give further insight into the mechanisms of gene regulation in apicomplexan parasites.
Post-transcriptional regulation
Whereas regulation of gene expression at the transcriptional level has been successful in a variety of organisms, the most favored technology in eukaryotes today is based on RNA interference (RNAi). First described in Caenorhabditis elegans (Fire et al. 1998), this technology has now revolutionized the field of molecular biology. However, the only protozoan parasite in which this tool has been substantially successful is Trypanosoma brucei. First described in 1998 (Ngo et al. 1998), it soon became the method of choice for generating conditional mutants for essential genes in this parasite [for recent reviews, see Motyka and Englund (2004); Clayton et al. (2005)].
Unfortunately, the RNAi pathway does not appear to be present in all protozoan parasites. Moreover, RNAi is not even conserved among members of the same phylum. For example, the machinery for RNAi appears to be absent from many close relatives of Trypanosoma brucei. Similarly, in other protozoa, the presence and efficiency of RNAi remains controversial. A good indication for a functional RNAi pathway is the presence of known RNAi-related genes, such as Argonaute (AGO-1), Dicer, or RNA-dependent RNA polymerase (RdRp; Ullu et al. 2004).
In E. histolytica, an RNAi pathway appears to be in place based on the presence of AGO-like proteins (Abed and Ankri 2005), and initial reports demonstrate the feasibility of using double-stranded RNA (dsRNA) to down regulate gene expression in this parasite (Kaur and Lohia 2004; Vayssie et al. 2004). In the case of G. lamblia and G. intestinalis, it has been reported that the machinery necessary for RNAi is present in the genomes (Ullu et al. 2004) and that small sense and antisense transcripts that might be employed in the RNAi pathway are also present (Ullu et al. 2005). Recently, a Dicer-like enzyme from G. intestinalis has been characterised in detail; it can produce small interfering RNAs (siRNAs) and is capable of functioning in the RNAi pathway of fission yeast (Macrae et al. 2006). Based on these findings, it can be anticipated that the RNAi pathway is indeed functional in these early-diverged eukaryotes and that RNAi as a molecular tool for the down-regulation of gene expression is possible to establish.
Although some reports suggest that RNAi appears operational in apicomplexan parasites, its study, thus far, has been restricted to only a few genes. The machinery for RNAi appears to be present in Toxoplasma gondii (Ullu et al. 2004; Al Riyahi et al. 2006), and some reports indicate that RNAi can down regulate gene expression (Al-Anouti and Ananvoranich 2002; Al-Anouti et al. 2004; Adams et al. 2005). In the case of Plasmodium species, multiple groups have failed to identify the components of the RNAi machinery. While some reports suggest that RNAi is functional in Plasmodium species (Malhotra et al. 2002; McRobert and McConkey 2002; Agrawal et al. 2003; Mohmmed et al. 2003), it cannot be ruled out at this point that the effects are non-specific. For example, treatment of parasites with dsRNA against P. falciparum falcipain-1 resulted in distinct morphological changes, including abnormally swollen food vacuoles (Malhotra et al. 2002). In contrast, ablation of the respective gene for falcipain-1 via homologous recombination had no effect on asexual growth, and no abnormally formed food vacuoles were observed (Eksi et al. 2004; Sijwali et al. 2004). Consequently, RNAi as a tool to diminish gene expression in apicomplexan parasites has not found widespread use to date.
The employment of antisense transcripts to down regulate gene expression has met with some success in protozoan parasites like G. lamblia (Davis-Hayman and Nash 2002) and Toxoplasma gondii (Nakaar et al. 1999). However, so far, the antisense effects observed are only partial, casting some limitations to this approach.
In summary, based on the biology of the respective parasite (life cycle, ploidy, genetic setup), different approaches to study the function of essential genes have been established. In the context of identifying novel candidates for vaccine or drug development, tools such as RNAi or antisense RNA may be adapted for high-throughput format to allow a relatively rapid analysis of gene function. Other technologies, such as the generation of conditional mutants using homologous recombination, are relatively laborious and would necessitate careful selection of target gene(s) to investigate. Considered with analyses of the genome and proteome, genetic and chemical screens may help to significantly narrow the list of candidates worthy of further investigation.
Forward genetic screens on parasites
Forward genetics, the generation of certain phenotypes and the subsequent identification of the gene, is a powerful strategy to identify novel genes (including hypothetical genes that are normally neglected in reverse genetic approaches). While technologies like RNAi in Trypanosoma brucei allow relatively easy gene-to-function studies in high-throughput format (see Motyka and Englund 2004), different approaches must be pursued to identify novel essential genes in forward genetic screens for other protozoan parasites. As mentioned above, the unique biology of each species has to be taken into consideration when designing a forward genetic screen. For example, insertional mutagenesis is a promising strategy to generate phenotypes in haploid organisms, but will prove difficult in diploids and tetraploids. Insertional mutagenesis can be performed relatively easily in systems where a powerful transfection system exists and non-homologous recombination occurs at reasonable frequency. In the following section, we will summarize a few strategies that are currently employed in apicomplexa and kinetoplastida.
Toxoplasma gondii grows rapidly in culture and is particularly amenable to genetic modification; consequently, much work has been done using this parasite to develop and enhance approaches to dissect gene function. In addition, several high-throughput assays have been established that permit the analysis of almost every step in the asexual life cycle. Apart from high-throughput growth assays using LacZ (Seeber and Boothroyd 1996; McFadden et al. 1997) or fluorescent markers (Gubbels and Striepen 2003), invasion and attachment of the host cell can be analyzed in laser scanning cytometer (LSC)-based assays (Mital et al. 2006). In the absence of reliable RNAi approaches, alternative strategies have been established to generate and identify parasites carrying mutations in critical genes. Initially, chemical mutagenesis was used to generate temperature-sensitive mutants (Pfefferkorn and Pfefferkorn 1976; Radke et al. 2000; Uyetake et al. 2001), allowing effective disruption of genes fulfilling critical roles during the life cycle of the parasite. The identification of the affected genes often proves difficult, however, because chemical mutagenesis may hit several independent genes. With the recent establishment of complementation systems for apicomplexan parasites (Striepen et al. 2002), the identification of single mutagenized genes appears to be possible via genetic rescue (White et al. 2005).
Another strategy broadly used in Toxoplasma gondii to generate a population of mutant parasites takes advantage of a highly efficient transfection system combined with random, non-homologous integration of the linear DNA transfected. Insertional mutagenesis has been used for the identification of promoters and tagging of genes with fluorescent proteins (Roos et al. 1997; Gubbels et al. 2004; Bradley et al. 2004). The technique has been particularly useful in the identification of genes involved in Toxoplasma gondii differentiation (Matrajt et al. 2002; Vanchinathan et al. 2005) or survival in activated macrophages (Mordue et al. 2007). Compared to chemical mutagenesis, the identification of the locus containing the insertion is relatively straightforward using plasmid rescue, inverse PCR, or other PCR-based approaches. The primary disadvantage of insertional mutagenesis is that only non-essential genes can be identified. In the future, it might be possible to combine the stringent conditional regulation provided by the Tet-inducible system (see above) with an insertional mutagenesis strategy (our unpublished work).
Exploring the phenome in Plasmodium species will no doubt be more complicated. While the establishment of a transposon-based transfection tool makes insertional mutagenesis appear possible (Balu et al. 2005; Balu and Adams 2006), the need for high-throughput analysis of the different steps involved in the life cycle of the parasite might complicate this approach. Another interesting systematic approach is a mutagenesis shuttle system, which allows identification of essential genes in a medium-throughput analysis (Sakamoto et al. 2005). Here, the transposon mutagenesis is performed in E. coli on large genomic fragments of P. berghei, which are then reintroduced into the parasite via homologous recombination. In the event a critical element is destroyed by the integration of the transposon, no homologous recombination can occur, and the subsequent failure to detect the corresponding event would indicate an essential function of the respective gene (Sakamoto et al. 2005).
Whereas the application of these strategies in haploid protozoa is efficient, in other protozoa, the diploid (or tetraploid) problem needs to be overcome (Beverley 2003), as two (or more) independent mutation events are required to result in a complete loss-of-function mutant. Although insertional mutagenesis for the generation of loss-of-function mutants is limited, it can be readily employed in these parasites to identify novel genes or promoter elements using trapping strategies. In particular, the use of transposons, like the mariner element of Drosophila melanogaster, has allowed researchers to adopt these strategies in kinetoplastida and other eukaryotes (Gueiros-Filho and Beverley 1997).
An alternative to genetic screens is the application of chemical genetics. In this study, chemical libraries are screened for an ability to block certain processes during the life cycle of the parasite. Upon identification of the respective compound, the challenge is then to identify the cellular target(s) [for a recent review, see Bogyo and Cravatt (2007)].
Conclusion
The study of cellular and molecular biology of protozoan parasites led to remarkable and surprising findings relevant for basic research and for the discovery of novel drug and vaccine candidates that are urgently needed to combat these pathogens in the future. As we enter into the post-genomic era, we are now faced with the longest list of putative candidates in need of functional analysis. Several remarkable tools to dissect gene function have been developed, but a new challenge awaits in adapting these tools for high or medium throughput analysis. The research community will benefit by keeping the current repertoire of tools in mind while brainstorming novel techniques to make molecular biological studies on protozoan parasites less arduous.
References
Abed M, Ankri S (2005) Molecular characterization of Entamoeba histolytica RNase III and AGO2, two RNA interference hallmark proteins. Exp Parasitol 110:265–269
Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V (2004) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445
Adams B, Musiyenko A, Kumar R, Barik S (2005) A novel class of dual-family immunophilins. J Biol Chem 280:24308–24314
Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK (2003) RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev 67:657–685
Al Riyahi A, Al-Anouti F, Al-Rayes M, Ananvoranich S (2006) Single argonaute protein from Toxoplasma gondii is involved in the double-stranded RNA induced gene silencing. Int J Parasitol 36:1003–1014
Al-Anouti F, Ananvoranich S (2002) Comparative analysis of antisense RNA, double-stranded RNA, and Delta ribozyme-mediated gene regulation in Toxoplasma gondii. Antisense nucleic acid drug dev 12:275–281
Al-Anouti F, Tomavo S, Parmley S, Ananvoranich S (2004) The expression of lactate dehydrogenase is important for the cell cycle of Toxoplasma gondii. J Biol Chem 279:52300–52311
Amino R, Menard R, Frischknecht F (2005) In vivo imaging of malaria parasites–recent advances and future directions. Curr Opin Microbiol 8:407–414
Arhets P, Olivo JC, Gounon P, Sansonetti P, Guillen N (1998) Virulence and functions of myosin II are inhibited by overexpression of light meromyosin in Entamoeba histolytica. Mol Biol Cell 9:1537–1547
Balu B, Adams JH (2006) Functional genomics of Plasmodium falciparum through transposon-mediated mutagenesis. Cell Microbiol 8:1529–1536
Balu B, Shoue DA, Fraser MJ Jr, Adams JH (2005) High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc Natl Acad Sci USA 102:16391–16396
Barrett B, LaCount DJ, Donelson JE (2004) Trypanosoma brucei: a first-generation CRE-loxP site-specific recombination system. Exp Parasitol 106:37–44
Beverley SM (2003) Protozomics: trypanosomatid parasite genetics comes of age. Nat Rev Genet 4:11–19
Bogyo M, Cravatt BF (2007) Genomics and proteomics from genes to function: advances in applications of chemical and systems biology. Curr Opin Chem Biol 11:1–3
Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1:e5
Bracha R, Nuchamowitz Y, Anbar M, Mirelman D (2006) Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog 2:e48
Bradley PJ, Li N, Boothroyd JC (2004) A GFP-based motif-trap reveals a novel mechanism of targeting for the Toxoplasma ROP4 protein. Mol Biochem Parasitol 137:111–120
Branda CS, Dymecki SM (2004) Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6:7–28
Brecht S, Erdhart H, Soete M, Soldati D (1999) Genome engineering of Toxoplasma gondii using the site-specific recombinase Cre. Gene 234:239–247
Brown M, Figge J, Hansen U, Wright C, Jeang KT, Khoury G, Livingston DM, Roberts TM (1987) Lac repressor can regulate expression from a hybrid SV40 early promoter containing a lac operator in animal cells. Cell 49:603–612
Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ (2002) Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512–519
Carvalho TG, Thiberge S, Sakamoto H, Menard R (2004) Conditional mutagenesis using site-specific recombination in Plasmodium berghei. Proc Natl Acad Sci USA 101:14931–14936
Clayton CE (1999) Genetic manipulation of kinetoplastida. Parasitol Today 15:372–378
Clayton C (2002) Life without transcriptional control? From fly to man and back again. EMBO 21:1881–1888
Clayton CE, Estevez AM, Hartmann C, Alibu VP, Field M, Horn D (2005) Down-regulating gene expression by RNA interference in Trypanosoma brucei. Methods Mol Biol 309:39–60
Cleary MD, Singh U, Blader IJ, Brewer JL, Boothroyd J (2002) Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot Cell 1:329–340
Corbel SY, Rossi FM (2002) Latest developments and in vivo use of the Tet system: ex vivo and in vivo delivery of tetracycline-regulated genes. Curr Opin Biotechnol 13:448–452
Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, Davern KM, Wickham ME, Brown GV, Coppel RL, Cowman AF (1997) Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell 89:287–296
Cui L, Fan Q,Li J (2002) The malaria parasite Plasmodium falciparum encodes members of the Puf RNA-binding protein family with conserved RNA binding activity. Nucleic Acids Res 30:4607–4617
Davis-Hayman SR, Nash TE (2002) Genetic manipulation of Giardia lamblia. Mol Biochem Parasitol 122:1–7
Deuschle U, Gentz R, Bujard H (1986) lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci USA 83:4134–4137
Eid J, Sollner-Webb B (1987) Efficient introduction of plasmid DNA into Trypanosoma brucei and transcription of a transfected chimeric gene. Proc Natl Acad Sci USA 84:7812–7816
Eksi S, Czesny B, Greenbaum DC, Bogyo M, Williamson KC (2004) Targeted disruption of Plasmodium falciparum cysteine protease, falcipain 1, reduces oocyst production, not erythrocytic stage growth. Mol Microbiol 53:243–250
Elmendorf HG, Singer SM, Nash TE (2001) The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res 29:4674–4683
El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B (2005a) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–415
El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, Ghedin E, Peacock C, Bartholomeu DC, Haas BJ, Tran AN, Wortman JR, Alsmark UC, Angiuoli S, Anupama A, Badger J, Bringaud F, Cadag E, Carlton JM, Cerqueira GC, Creasy T, Delcher AL, Djikeng A, Embley TM, Hauser C, Ivens AC, Kummerfeld SK, Pereira-Leal JB, Nilsson D, Peterson J, Salzberg SL, Shallom J, Silva JC, Sundaram J, Westenberger S, White O, Melville SE, Donelson JE, Andersson B, Stuart KD, Hall N (2005b) Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404–409
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Gale M Jr, Carter V, Parsons M (1994) Translational control mediates the developmental regulation of the Trypanosoma brucei Nrk protein kinase. J Biol Chem 269:31659–31665
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan M-S, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511
Gardner MJ, Bishop R, Shah T, de Villiers EP, Carlton JM, Hall N, Ren Q, Paulsen IT, Pain A, Berriman M, Wilson RJ, Sato S, Ralph SA, Mann DJ, Xiong Z, Shallom SJ, Weidman J, Jiang L, Lynn J, Weaver B, Shoaibi A, Domingo AR, Wasawo D, Crabtree J, Wortman JR, Haas B, Angiuoli SV, Creasy TH, Lu C, Suh B, Silva JC, Utterback TR, Feldblyum TV, Pertea M, Allen J, Nierman WC, Taracha EL, Salzberg SL, White OR, Fitzhugh HA, Morzaria S, Venter JC, Fraser CM, Nene V (2005) Genome sequence of Theileria parva, a bovine pathogen that transforms lymphocytes. Science 309:134–137
Gatz C, Quail PH (1988) Tn10-encoded tet repressor can regulate an operator-containing plant promoter. Proc Natl Acad Sci USA 85:1394–1397
Gatz C, Frohberg C, Wendenburg R (1992) Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J 2:397–404
Goodrich JA, Kugel JF (2006) Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol 7:612–616
Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89:5547–5551
Gubbels MJ, Striepen B (2003) High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob Agents Chemother 47:309–316
Gubbels MJ, Striepen B (2004) Studying the cell biology of apicomplexan parasites using fluorescent proteins. Microsc Microanal 10:568–579
Gubbels MJ, Wieffer M, Striepen B (2004) Fluorescent protein tagging in Toxoplasma gondii: identification of a novel inner membrane complex component conserved among Apicomplexa. Mol Biochem Parasitol 137:99–110
Gueiros-Filho FJ, Beverley SM (1997) Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276:1716–1719
Gunasekera AM, Patankar S, Schug J, Eisen G, Kissinger J, Roos D, Wirth DF (2004) Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol Biochem Parasitol 136:35–42
Hamann L, Buss H, Tannich E (1997) Tetracycline-controlled gene expression in Entamoeba histolytica. Mol Biochem Parasitol 84:83–91
Hariharan S, Ajioka J, Swindle J (1993) Stable transformation of Trypanosoma cruzi: inactivation of the PUB12.5 polyubiquitin gene by targeted gene disruption. Mol Biochem Parasitol 57:15–30
Hinnen A, Hicks JB, Fink GR (1978) Transformation of yeast. Proc Natl Acad Sci USA 75:1929–1933
Hu MC, Davidson N (1987) The inducible lac operator–repressor system is functional in mammalian cells. Cell 48:555–566
Huynh MH, Carruthers VB (2006) Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog 2(8):e84
Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O’Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442
Kaur G, Lohia A (2004) Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem Biophys Res Commun 320:1118–1122
Kim K, Soldati D, Boothroyd JC (1993) Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science 262:911–914
Kooij TW, Carlton JM, Bidwell SL, Hall N, Ramesar J, Janse CJ, Waters AP (2005) A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog 1:e44
Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De la Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503–1508
Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillen N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri Jr WA, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N (2005) The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868
Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311:195–198
Mahner M, Kary M (1997) What exactly are genomes, genotypes and phenotypes? And what about phenomes? J Theor Biol 186:55–63
Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, Khan SM, Dimopoulos G, Janse CJ, Waters AP (2006) Regulation of sexual development of Plasmodium by translational repression. Science 313:667–669
Malhotra P, Dasaradhi PV, Kumar A, Mohmmed A, Agrawal N, Bhatnagar RK, Chauhan VS (2002) Double-stranded RNA-mediated gene silencing of cysteine proteases (falcipain-1 and -2) of Plasmodium falciparum. Mol Microbiol 45:1245–1254
Matrajt M, Donald RG, Singh U, Roos DS (2002) Identification and characterization of differentiation mutants in the protozoan parasite Toxoplasma gondii. Mol Microbiol 44:735–747
Mazumdar J, Wilson EH, Masek K, Hunter CA, Striepen B (2006) Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc Natl Acad Sci USA 103:13192–13197
McFadden DC, Seeber F, Boothroyd JC (1997) Use of Toxoplasma gondii expressing beta-galactosidase for colorimetric assessment of drug activity in vitro. Antimicrob Agents Chemother 41:1849–1853
McRobert L, McConkey GA (2002) RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol Biochem Parasitol 119:273–278
Meissner M, Soldati D (2005) The transcription machinery and the molecular toolbox to control gene expression in Toxoplasma gondii and other protozoan parasites. Microbes Infect 7:1376–1384
Meissner M, Brecht S, Bujard H, Soldati D (2001) Modulation of myosin A expression by a newly established tetracycline repressor-based inducible system in Toxoplasma gondii. Nucleic Acids Res 29:E115
Meissner M, Schluter D, Soldati D (2002) Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298:837–840
Meissner M, Krejany E, Gilson PR, de Koning-Ward TF, Soldati D, Crabb BS (2005) Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci USA 102:2980–2985
Metzger D, Clifford J, Chiba H, Chambon P (1995) Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA 92:6991–6995
Mirelman D, Anbar M, Nuchamowitz Y, Bracha R (2006) Epigenetic silencing of gene expression in Entamoeba histolytica. Arch Med Res 37:226–233
Mital J, Meissner M, Soldati D, Ward GE (2005) Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol Biol Cell 16:4341–4349
Mital J, Schwarz J, Taatjes DJ, Ward GE (2006) Laser scanning cytometer-based assays for measuring host cell attachment and invasion by the human pathogen Toxoplasma gondii. Cytometry A 69:13–19
Mohmmed A, Dasaradhi PV, Bhatnagar RK, Chauhan VS, Malhotra P (2003) In vivo gene silencing in Plasmodium berghei—a mouse malaria model. Biochem Biophys Res Commun 309:506–511
Mordue DG, Scott-Weathers CF, Tobin CM, Knoll LJ (2007) A patatin-like protein protects Toxoplasma gondii from degradation in activated macrophages. Mol Microbiol 63:482–496
Motyka SA, Englund PT (2004) RNA interference for analysis of gene function in trypanosomatids. Curr Opin Microbiol 7:362–368
Nakaar V, Samuel BU, Ngo EO, Joiner KA (1999) Targeted reduction of nucleoside triphosphate hydrolase by antisense RNA inhibits Toxoplasma gondii proliferation. J Biol Chem 274:5083–5087
Ngo H, Tschudi C, Gull K, Ullu E (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA 95:14687–14692
Nickel R, Tannich E (1994) Transfection and transient expression of chloramphenicol acetyltransferase gene in the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci USA 91:7095–7098
Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR Jr, Fidock DA (2006) Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods 3:615–621
Pain A, Renauld H, Berriman M, Murphy L, Yeats CA, Weir W, Kerhornou A, Aslett M, Bishop R, Bouchier C, Cochet M, Coulson RM, Cronin A, de Villiers EP, Fraser A, Fosker N, Gardner M, Goble A, Griffiths-Jones S, Harris DE, Katzer F, Larke N, Lord A, Maser P, McKellar S, Mooney P, Morton F, Nene V, O’Neil S, Price C, Quail MA, Rabbinowitsch E, Rawlings ND, Rutter S, Saunders D, Seeger K, Shah T, Squares R, Squares S, Tivey A, Walker AR, Woodward J, Dobbelaere DA, Langsley G, Rajandream MA, McKeever D, Shiels B, Tait A, Barrell B, Hall N (2005) Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science 309:131–133
Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF (2001) Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol Biol Cell 12:3114–3125
Pfefferkorn ER, Pfefferkorn LC (1976) Toxoplasma gondii: isolation and preliminary characterization of temperature-sensitive mutants. Exp Parasitol 39:365–376
Purdy JE, Mann BJ, Pho LT, Petri WA Jr, (1994) Transient transfection of the enteric parasite Entamoeba histolytica and expression of firefly luciferase. Proc Natl Acad Sci USA 91:7099–7103
Radke JR, Guerini MN, White MW (2000) Toxoplasma gondii: characterization of temperature-sensitive tachyzoite cell cycle mutants. Exp Parasitol 96:168–177
Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW (2005) The transcriptome of Toxoplasma gondii. BMC Biol 3:26
Ramakrishnan G, Vines RR, Mann BJ, Petri Jr WA (1997) A tetracycline-inducible gene expression system in Entamoeba histolytica. Mol Biochem Parasitol 84:93–100
Roos DS, Sullivan WJ, Striepen B, Bohne W, Donald RG (1997) Tagging genes and trapping promoters in Toxoplasma gondii by insertional mutagenesis. Methods 13:112–122
Sahoo N, Bhattacharya S, Bhattacharya A (2003) Blocking the expression of a calcium binding protein of the protozoan parasite Entamoeba histolytica by tetracycline regulatable antisense-RNA. Mol Biochem Parasitol 126:281–284
Sakamoto H, Thiberge S, Akerman S, Janse CJ, Carvalho TG, Menard R (2005) Towards systematic identification of Plasmodium essential genes by transposon shuttle mutagenesis. Nucleic Acids Res 33:e174
Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ Jr, Cesbron-Delauw MF, Hakimi MA (2005) Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol 25:10301–10314
Seeber F, Boothroyd JC (1996) Escherichia coli beta-galactosidase as an in vitro and in vivo reporter enzyme and stable transfection marker in the intracellular protozoan parasite Toxoplasma gondii. Gene 169:39–45
Sijwali PS, Kato K, Seydel KB, Gut J, Lehman J, Klemba M, Goldberg DE, Miller LH, Rosenthal PJ (2004) Plasmodium falciparum cysteine protease falcipain-1 is not essential in erythrocytic stage malaria parasites. Proc Natl Acad Sci USA 101:8721–8726
Simpson L, Sbicego S, Aphasizhev R (2003) Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9:265–276
Soldati D, Boothroyd JC (1993) Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260:349–352
Soldati D, Boothroyd JC (1995) A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol 15:87–93
Striepen B, White MW, Li C, Guerini MN, Malik SB, Logsdon JM Jr, Liu C, Abrahamsen MS (2002) Genetic complementation in apicomplexan parasites. Proc Natl Acad Sci USA 99:6304–6309
Sullivan WJ Jr, Narasimhan J, Bhatti MM, Wek RC (2004) Parasite-specific eIF2 (eukaryotic initiation factor-2) kinase required for stress-induced translation control. Biochem J 380:523–531
Sullivan WJ Jr, Naguleswaran A, Angel SO (2006) Histones and histone modifications in protozoan parasites. Cell Microbiol 8:1850–1861
Sun CH, Tai JH (2000) Development of a tetracycline controlled gene expression system in the parasitic protozoan Giardia lamblia. Mol Biochem Parasitol 105:51–60
Teixeira SM, daRocha WD (2003) Control of gene expression and genetic manipulation in the Trypanosomatidae. Genet Mol Res 2:148–158
Touz MC, Conrad JT, Nash TE (2005) A novel palmitoyl acyl transferase controls surface protein palmitoylation and cytotoxicity in Giardia lamblia. Mol Microbiol 58:999–1011
Ullu E, Tschudi C, Chakraborty T (2004) RNA interference in protozoan parasites. Cell Microbiol 6:509–519
Ullu E, Lujan HD, Tschudi C (2005) Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell 4:1155–1157
Uyetake L, Ortega-Barria E, Boothroyd JC (2001) Isolation and characterization of a cold-sensitive attachment/invasion mutant of Toxoplasma gondii. Exp Parasitol 97:55–59
van Dijk MR, Waters AP, Janse CJ (1995) Stable transfection of malaria parasite blood stages. Science 268:1358–1362
van Noort V, Huynen MA (2006) Combinatorial gene regulation in Plasmodium falciparum. Trends Genet 22:73–78
van Poppel NF, Welagen J, Duisters RF, Vermeulen AN, Schaap D (2006) Tight control of transcription in Toxoplasma gondii using an alternative tet repressor. Int J Parasitol 36:443–452
Vanacova S, Liston DR, Tachezy J, Johnson PJ (2003) Molecular biology of the amitochondriate parasites, Giardia intestinalis, Entamoeba histolytica and Trichomonas vaginalis. Int J Parasitol 33:235–255
Vanchinathan P, Brewer JL, Harb OS, Boothroyd JC, Singh U (2005) Disruption of a locus encoding a nucleolar zinc finger protein decreases tachyzoite-to-bradyzoite differentiation in Toxoplasma gondii. Infect Immun 73:6680–6688
Vats D, Vishwakarma RA, Bhattacharya S, Bhattacharya A (2005) Reduction of cell surface glycosylphosphatidylinositol conjugates in Entamoeba histolytica by antisense blocking of E. histolytica GlcNAc-phosphatidylinositol deacetylase expression: effect on cell proliferation, endocytosis, and adhesion to target cells. Infect Immun 73:8381–8392
Vayssie L, Vargas M, Weber C, Guillen N (2004) Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol Biochem Parasitol 138:21–28
Waller RF, McFadden GI (2005) The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol 7:57–79
Wang Z, Morris JC, Drew ME, Englund PT (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem 275:40174–40179
White MW, Jerome ME, Vaishnava S, Guerini M, Behnke M, Striepen B (2005) Genetic rescue of a Toxoplasma gondii conditional cell cycle mutant. Mol Microbiol 55:1060–1071
Wirtz E, Clayton C (1995) Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268:1179–1183
Wirtz E, Hoek M, Cross GA (1998) Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res 26:4626–4634
Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE (1995) Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci USA 92:973–977
Wu Y, Kirkman LA, Wellems TE (1996) Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc Natl Acad Sci USA 93:1130–1134
Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA (2004) The genome of Cryptosporidium hominis. Nature 431:1107–1112
Yan S, Myler PJ, Stuart KD (2001) Tetracycline regulated gene expression in Leishmania donovani. Mol Biochem Parasitol 112:61–69
Yee J, Nash TE (1995) Transient transfection and expression of firefly luciferase in Giardia lamblia. Proc Natl Acad Sci USA 92:5615–5619
Acknowledgment
We thank all the investigators who are investing time on tool development in protozoa for the recent advances, some of which we were not able to mention due to space restrictions. We are indebted to Dr. Friedrich Frischknecht and Dr. Angelika Herm-Götz for carefully reading the manuscript. M.M. is funded by the BioFuture programme of the BMBF, C.A.-N. is funded by the Schwerpunktprogramm 688 of the DFG. The research of W.J.S. is supported by the Public Health Service grant GM065051 from the National Institutes of Health and the American Heart Association (0750201Z).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meissner, M., Agop-Nersesian, C. & Sullivan, W.J. Molecular tools for analysis of gene function in parasitic microorganisms. Appl Microbiol Biotechnol 75, 963–975 (2007). https://doi.org/10.1007/s00253-007-0946-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-0946-4