Abstract
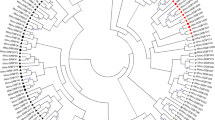
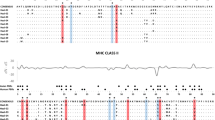
The major histocompatibility complex (MHC) is a highly variable family of genes involved in parasite recognition and the initiation of adaptive immune system responses. Variation in MHC loci is maintained primarily through parasite-mediated selection or disassortative mate choice. To characterize MHC diversity of rufous-collared sparrows (Zonotrichia capensis), an abundant South American passerine, we examined allelic and nucleotide variation in MHC class I exon 3 using pyrosequencing. Exon 3 comprises a substantial portion of the peptide-binding region (PBR) of class I MHC and thus plays an important role in intracellular pathogen defense. We identified 98 putatively functional alleles that produce 56 unique protein sequences across at least 6 paralogous loci. Allelic diversity per individual and exon-wide nucleotide diversity were relatively low; however, we found specific amino acid positions with high nucleotide diversity and signatures of positive selection (elevated d N /d S ) that may correspond to the PBR. Based on the variation in physicochemical properties of amino acids at these “positively selected sites,” we identified ten functional MHC supertypes. Spatial variation in nucleotide diversity and the number of MHC alleles, proteins, and supertypes per individual suggests that environmental heterogeneity may affect patterns of MHC diversity. Furthermore, populations with high MHC diversity have higher prevalence of avian malaria, consistent with parasite-mediated selection on MHC. Together, these results provide a framework for subsequent investigations of selective agents acting on MHC in Z. capensis.




Similar content being viewed by others
References
Alcaide M, Edwards SV, Negro JJ (2007) Characterization, polymorphism, and evolution of MHC class II B genes in birds of prey. J Mol Evol 65:541–554
Alcaide M, Edwards SV, Cadahía L, Negro JJ (2009) MHC class I genes of birds of prey: isolation, polymorphism and diversifying selection. Conserv Genet 10:1349–1355
Alcaide M, Lui M, Edward SV (2013) Major histocompatibility complex class I evolution in songbirds: universal primers, rapid evolution and base compositional shifts in exon 3. Peer J 1:e86
Anisimova M, Nielsen R, Yang Z (2003) Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229–1236
Babik W, Pabijan M, Arntzen JW, Cogălniceanu D, Durka W, Radwan J (2009a) Long-term survival of a urodele amphibian despite depleted major histocompatibility complex variation. Mol Ecol 18:769–781
Babik W, Taberlet P, Ejsmond MJ, Radwan J (2009b) Next generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol Ecol Res 9:713–719
Babik W, Kawałko A, Wójcik JM, Radwan J (2012) Low major histocompatibility complex class I (MHC I) variation in the European Bison (Bison bonasus). J Hered 103:349–359
Balakrishnan CN, Ekblom R, Völker M, Westerdahl H, Godinez R, Kotkiewicz H, Burt DW, Graves T, Griffin DK, Warren WC, Edwards SV (2010) Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biol 8:29
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16:363–377
Bjorkman PJ, Saper MA, Samraoui BSB, Bennett WS, Strominger JL, Wiley DC (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512–518
Bonneaud C, Sorci G, Morin V, Westerdahl H, Zoorob R, Wittzell H (2004) Diversity of Mhc class I and IIB genes in house sparrows (Passer domesticus). Immunogenetics 55:855–865
Bonneaud C, Pérez-Tris J, Federici P, Chastel O, Sorci G (2006) Major histocompatibility alleles associated with local resistance to malaria in a passerine. Evolution 60:383–389
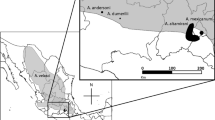
Cheviron ZA, Brumfield RT (2009) Migration-selection balance and local adaptation of mitochondrial haplotypes in rufous-collared sparrow (Zonotrichia capensis) along an elevational gradient. Evolution 63:1593–1605
Cole RK (1968) Studies on the genetic resistance to Marek’s disease. Avian Dis 12:9–28
Delany ME, Robinson CM, Goto RM, Miller MM (2009) Architecture and organization of chicken microchromosome 16: order of the NOR, MHC-Y, and MHC-B subregions. J Hered 100:507–514
Ditchkoff SS, Lochmiller RL, Masters RE, Hoofer SR, Van Den Bussche RA (2001) Major histocompatibility-complex-associated variation in secondary sexual traits of white tailed deer (Odocoileus virginianus): evidence for good-genes advertisement. Evolution 55:616–625
Doytchinova IA, Flower DR (2005) In silico identification of supertypes for class II MHCs. J Immunol 174:7085–7095
Dunn PO, Bollmer JL, Freeman-Gallant CR, Whittingham LA (2013) MHC variation is related to a sexually selected ornament, survival, and parasite resistance in common yellowthroats. Evolution 67:679–687
Edwards SV, Hedrick PW (1998) Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol Evol 13:305–311
Ekblom R, Stapley J, Ball AD, Birkhead T, Burke T, Slate J (2011) Genetic mapping of the major histocompatibility complex in the zebra finch (Taeniopygia guttata). Immunogenetics 63:523–530
Ellison A, Allainguillaume J, Girdwood S, Pachebat J, Peat KM, Wright P, Consuegra S (2012) Maintaining functional major histocompatibility complex diversity under inbreeding: the case of a selfing vertebrate. Proc R Soc B 279:5004–5013
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481–501
Galan M, Guivier E, Caraux G, Charbonnel N, Cosson J-F (2010) A 454 multiplex sequencing method for rapid and reliable genotyping of highly polymorphic genes in large-scale studies. BMC Genomics 11:296
Gilles A, Meglécz E, Pech N, Ferreira S, Malausa T, Martin JF (2011) Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 12:245
Hackett SJ et al (2008) A phylogenomic study of birds reveals their evolutionary history. Science 320:1763–1768
Hammer J, Gallazzi F, Bono E, Karr RW, Guenot J, Valsasnini J, Nagy ZA, Sinigaglia FL (1995) Peptide binding specificity of HLA-DR4 molecules: correlation with rheumatoid arthritis association. J Exp Med 5:1847–1855
Hansen MM, Skaala O, Jensen LF, Bekkevold D, Mensberg KL (2007) Gene flow, effective population size and selection at major histocompatibility complex genes: brown trout in the Hardanger Fjord, Norway. Mol Ecol 16:1413–1425
Hedrick PW (2002) Pathogen resistance and genetic variation at MHC loci. Evolution 56(1902):1908
Hill AVS (1998) The immunogenetics of human infectious diseases. Ann Rev Immunol 16:593–617
Horton R, Wilming L, Rand V, Lovering RC, Bruford AE, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 5:889–899
Hughes AL, Nei M (1988) Patterns of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170
Hughes AL, Nei M (1989) Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol Biol Evol 6:559–579
Hughes CR, Miles S, Walbroehl JM (2008) Support for the minimal essential MHC hypothesis: a parrot with a single, highly polymorphic MHC class II B gene. Immunogenetics 60:219–231
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Jones MR, Cheviron ZA, Carling MD (2013) Spatial patterns of avian malaria prevalence in Zonotrichia capensis on the western slope of the Peruvian Andes. J Parasitol 99:903–905
Kaufman J, Milne S, Göbel TWF, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 40:923–925
Klein J, Satta Y, O’hUigin C, Takahata N (1993) The molecular descent of the major histocompatibility complex. Ann Rev Immunol 11:269–295
Kloch A, Babik W, Bajer A, Siński A, Radwan J (2010) Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol Ecol 19:255–265
Kryazhimskiy S, Plotkin JB (2008) The population genetics of dN/dS. PLoS Genet 4:e1000304
Lamaze FC, Pavey SA, Normandeau E, Roy G, Garant D, Bernatchez L (2014) Natural and selective processes shape MHC gene diversity and expression in stocked brook charr populations (Salvelinus fontinalis). Mol Ecol 23:1730–1748
LePage KT, Miller MM, Briles WE, Taylor RL Jr (2000) Rfp-Y genotype affects the fate of Rous sarcomas in B2 B5 chickens. Immunogenetics 51:751–754
Loiseau C, Zoorob R, Robert A, Chastel O, Julliard R, Sorci G (2010) Plasmodium relictum infection and MHC diversity in the House Sparrow. Proc R Soc B 278:1264–1272
Lougheed SC, Campagna L, Dávila JA, Tubaro PL, Lijtmaer DA, Handford P (2013) Continental phylogeography of an ecologically and morphologically diverse songbird, Zonotrichia capensis. BMC Evol Biol 13:58
Matsumura M, Fremont DH, Peterson PA, Wilson IA (1992) Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science 257:927–934
Miller HC, Bowker-Wright G, Kharkrang M, Ramstad K (2011) Characterisation of class II B MHC genes from a ratite bird, the little spotted kiwi (Apteryx owenii). Immunogenetics 63:223–233
Møller AP, Erritzøe J (1998) Host immune defence and migration in birds. Evol Ecol 12:945–953
Nadachowska-Brzyska K, Zieliński P, Radwan J, Babik W (2012) Interspecific hybridization increases MHC class II diversity in two sister species of newts. Mol Ecol 21:887–906
Ophir R, Graur D (1997) Patterns and rates of indel evolution in processed pseudogenes from humans and murids. Gene 205:191–202
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420
Penn DJ, Damjanovich K, Potts WK (2002) MHC heterozygosity confers a selective advantage against multiple strain infections. Proc Natl Acad Sci 99:11260–11264
Promerová M, Babik W, Bryja J, Albrecht T, Stuglik M, Radwan J (2012) Evaluation of two approaches to genotyping major histocompatibility complex class I in passerine CE SSCP and 454 pyrosequencing. Mol Ecol Res 12:285–292
R Development Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Reusch TBH, Häberli MA, Aeschlimann PB, Milinski M (2001) Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature 414:300–302
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Schulenberg TS, Stotz DF, Lane DF, O’Neill JP, Parker TA III (2007) Birds of Peru. Princeton University Press, Princeton, p 656
Schut E, Rivero-de Aguilar J, Merino S, Magrath MJL, Komdeur J, Westerdahl H (2011) Characterization of MHC-I in the blue tit (Cyanistes caeruleus) reveals low levels of genetic diversity and trans-population evolution across European populations. Immunogenetics 63:531–542
Sepil I, Moghadam HK, Huchard E, Sheldon BC (2012) Characterization and 45 pyrosequencing of major histocompatibility complex class I genes in the great tit reveal complexity in a passerine system. BMC Evol Biol 12:68
Shetty S, Griffin DK, Graves JA (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosom Res 7:289–295
Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, Hanzawa K, Beck S, Kulski JK, Inoko H (2004) Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 172:6751–6763
Shiina T, Briles WE, Goto RM, Hosomichi K, Yanagiya K, Shimizu S, Inoko H, Miller MM (2007) Extended gene map reveals tripartite motif, C-type lectin, and Ig superfamily type genes within a subregion of the chicken MHC-B affecting infectious disease. J Immunol 178:7162–7172
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms, and misunderstandings. Proc R Soc B 277:979–988
Spurgin LG, van Oosterhout C, Illera JC, Bridgett S, Gharbi K, Emerson BC, Richardson DS (2011) Gene conversion rapidly generates major histocompatibility complex diversity in recently founded bird populations. Mol Ecol 20:5213–5225
Stager M, Cerasale DJ, Dor R, Winkler DW, Cheviron ZA (2014) Signatures of natural selection in the mitochondrial genomes of Tachycineta swallows and their implications for latitudinal patterns of the ‘pace of life’. Gene. In press.
Stiebens VA, Merino SE, Chain FJJ, Eizaguirre C (2013) Evolution of MHC class I genes in the endangered loggerhead sea turtle (Caretta caretta) revealed by 454 amplicon sequencing. BMC Evol Biol 13:95
Strandh M, Lannefors M, Bonadonna F, Westerdahl H (2011) Characterization of MHC class I and II genes in a subantarctic seabird, the blue petrel, Halobaena caerulea (Procellariiformes). Immunogenetics 63:653–666
Strandh M, Westerdahl H, Pontarp M, Canbäck B, Dubois MP, Miquel C, Taberlet P, Bonadonna F (2012) Major histocompatibility complex class II compatibility, but not class I, predicts mate choice in a bird with highly developed olfaction. Proc R Soc B 279:4457–4463
Stuglik MT, Radwan J, Babik W (2011) jMHC: software assistant for multilocus genotyping of gene families using next-generation amplicon sequencing. Mol Ecol Res 11:739–742
Sutton JT, Nakagawa S, Robertson BC, Jamieson IG (2011) Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Mol Ecol 20:4408–4420
Sutton JT, Robertson BC, Grueber CE, Stanton JL, Jamieson IG (2013) Characterization of MHC class II B polymorphism in bottlenecked New Zealand saddlebacks reveals low levels of genetic diversity. Immunogenetics 65:619–633
Takahashi K, Rooney AP, Nei M (2000) Origins and divergence times of mammalian class II MHC gene clusters. J Hered 91:198–204
Takahata N, Nei M (1990) Allelic genealogy under overdominant and frequency dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124:967–978
Tao N, Bruno WJ, Abfalterer W, Moret BME, Leitner, T, Kuiken C (2005) FINDMODEL: a tool to select the best-fit model of nucleotide substitution. http://hcv.lanl.gov/content/sequence/findmodel/findmodel.html
von-Schantz T, Wittzell H, Göransson G, Grahn M, Persson K (1996) MHC genotype and male ornamentation: genetic evidence for the Hamilton-Zuk hypothesis. Proc R Soc B 263:265–271
Wakenell PS, Miller MM, Goto RM, Gauderman WJ, Briles WE (1996) Association between the Rfp-Y haplotype and the incidence of Marek’s disease in chickens. Immunogenetics 44:242–245
Wallny H-J, Avila D, Hunt LG, Powell TJ, Riegert P, Salomonsen J, Skjødt K, Vainio O, Vilbois F, Wiles MV, Kaufman J (2006) Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci 103:1434–1439
Wang Y, Qiu M, Yang J, Zhao X, Wang Y, Zhu Q, Liu Y (2014) Sequence variations of the MHC class I gene exon 2 and exon 3 between infected and uninfected chicken challenged with Marek’s disease virus. Infect Genet Evol 21:103–109
Wegner KM, Reusch TBH, Kalbe M (2003) Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J Evol Biol 16:224–232
Westerdahl H, Wittzell H, von-Schantz T (1999) Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics 49:158–170
Westerdahl H, Waldenström J, Hansson B, Hasselquist D, von-Schantz T, Bensch S (2005) Associations between malaria and MHC genes in a migratory songbird. Proc R Soc B 272:151–1518
Westerdahl H (2007) Passerine MHC: genetic variation and disease resistance in the wild. J Ornithol 148:S469–S477
Westerdahl H, Asghar M, Hasselquist D, Bensch S (2012) Quantitative disease resistance: to better understand parasite-mediated selection on major histocompatibility complex. Proc R Soc B 279:577–584
Wilson DJ, McVean G (2006) Estimating diversifying selection and functional constraint in the presence of recombination. Genetics 172:1411–1425
Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E Jr, Wright C, Harkins T, O’Connor DH (2009) Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 15:1322–1327
Wucherpfennig KW, Yu B, Bhol K, Monos DS, Argyris E, Karr RW, Ahmed AR, Strominger JL (1995) Structural basis for major histocompatibility complex (MHC)-linked susceptibility to autoimmunity: charged residues of a single MHC binding pocket confer selective presentation of self-peptides in pemphigus vulgaris. Proc Natl Acad Sci 92:11935–11939
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Swanson WJ (2002) Codon-substitution models to detect adaptive evolution that account for heterogenous selective pressures among site classes. Mol Biol Evol 19:49–57
Zagalska-Neubauer M, Babik W, Stuglik M, Gustafsson L, Cichoń M, Radwan J (2010) 454 sequencing reveals extreme complexity of the class II major histocompatibility complex in the collared flycatcher. BMC Evol Biol 10:395
Acknowledgments
This work was funded by the American Museum of Natural History Frank M. Chapman Fund, Sigma Xi Grant-In-Aid-Of-Research, the American Ornithologists’ Union, and the University of Wyoming. Voucher specimens and pectoral muscle tissue samples of all of the specimens included in this study are accessioned at the Louisiana State University Museum of Natural Science (Baton Rouge), the Museo de Historia Natural, Universidad Nacional Mayor de San Marcos (Lima, Peru), and the Centro de Ornitologia y Biodiversidad (Lima, Peru). The Genome Sequencing and Analysis Core Resource at Duke University sequenced MHC amplicon libraries. We thank Amy Ellison for advice with designing MHC sequencing protocol. We thank Shawn M. Billerman, C. Alex Buerkle, Michael E. Dillon, James M. Maley, Melanie M. Murphy, and three anonymous reviewers for providing helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jones, M.R., Cheviron, Z.A. & Carling, M.D. Variation in positively selected major histocompatibility complex class I loci in rufous-collared sparrows (Zonotrichia capensis). Immunogenetics 66, 693–704 (2014). https://doi.org/10.1007/s00251-014-0800-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-014-0800-7