Abstract
Single nucleotide polymorphisms (SNPs) in the proximity of the interleukin-28B (IL28B) gene can predict spontaneous resolution of hepatitis C virus (HCV) infection and response to interferon therapy. Screening for this polymorphism has become part of the standard criteria for the management of HCV-infected patients, hence the need for a rapid, cost-effective screening method. Here, we describe a rapid PCR-based test to screen for two IL28B SNPs (rs12979860 and rs8099917). We used this test to investigate IL28B polymorphism and prevalence in a cohort of French Canadian injection drug users who are part of a unique population known to have a strong genetic founder effect. This population had lower linkage disequilibrium between the two tested SNPs as compared to other cohorts (|d′| = 0.68, r = 0.59). The special genetic makeup should be considered in the management of HCV-infected patients within that population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis C virus (HCV) infection is a major health problem affecting 130 million individuals worldwide with an increasing rate of new infections among injection drug users (IDUs) (Alter 2007). While approximately 25 % of HCV-exposed individuals can clear the virus spontaneously, the majority become persistently infected (Hoofnagle 2002). Chronic HCV infection leads to progressive liver damage over a period of years or decades and even liver cancer (Heathcote 2004; Hoofnagle 2002). Several studies in humans and chimpanzees have demonstrated that adaptive T cell responses are essential for spontaneous viral clearance (Dustin and Rice 2007; Shoukry et al. 2004; Walker 2010) but little is known about host genetic factors that influence HCV resolution.
Recent genome-wide association studies (GWAS) have identified polymorphism in or around the interleukin-28B (IL28B) gene as an important predictor of therapeutic and spontaneous HCV clearance (Ge et al. 2009; Suppiah et al. 2009; Thomas et al. 2009). The correlation with therapeutic HCV clearance was validated by several groups and across different genotypes (Mangia et al. 2010) as well as in individuals coinfected with human immunodeficiency virus (HIV) (Rallon et al. 2010; Rauch et al. 2010). Two main single nucleotide polymorphisms (SNPs) were identified as the major predictors of viral clearance, known as sustained viral response, following interferon (IFN) therapy: rs12979860 (3 kb upstream of IL28B) and rs8099917 (8 kb upstream of IL28B). Given that the IL28B and IL29 genes are in opposite orientation, these SNPs are upstream of both genes. It was also demonstrated that these two SNPs are in linkage disequilibrium. For rs12979860, the responder genotype is identified as C and the nonresponder as T. For rs8099917, the responder genotype is identified as T and the nonresponder genotype as G. The responder genotypes were linked to a higher expression of IL28A/B as measured by qRT-PCR in peripheral blood mononuclear cells (PBMCs) (Suppiah et al. 2009) and lower HCV viral load. The nonresponder genotypes were associated with higher expression of IFN-stimulated gene signal in the liver (Ge et al. 2009), which is considered as a predictor of nonresponse to IFN therapy (Chen et al. 2005). The rs12979860 SNP was also strongly associated with spontaneous viral clearance (Rauch et al. 2010; Thomas et al. 2009; Thompson et al. 2010). The predictive value was even higher when combined with plasma levels of interferon-γ-inducible protein-10 (IP-10) in the context of response to therapy (Darling et al. 2011) and spontaneous resolution (Beinhardt et al. 2012).
IL28B genotyping has become an important diagnostic tool for the management of HCV-infected patients. This test will continue to be important for new treatment regimens using direct acting antivirals in combination with IFN (Holmes et al. 2012) and even in IFN-free regimens (Barreiro et al. 2012). While a number of tests based on RT-PCR and direct sequencing were developed and are now available commercially, there is a need for a rapid and inexpensive test especially in a research or limited resources setting. The goals of this study were as follows: first, to develop a rapid, PCR-based technique for screening of both rs12979860 and rs8099917 and, second, to use this validated method to evaluate the prevalence of the different IL28B SNPs in a cohort of IDUs at high risk of HCV infection in Montréal, QC, Canada. This cohort is mainly composed of French Canadians, a population known to have a strong founder effect (Roy-Gagnon et al. 2011; Scriver 2001) which may influence the SNP distribution and consequently clinical management of this cohort.
Patients and methods
Study population
Participants were recruited among high-risk IDUs, defined as IDUs who reported sharing injection material or having an HCV-positive partner in the past 6 months, participating in the Montreal Acute Hepatitis C Cohort (HEPCO) at St-Luc Hospital of the Centre Hospitalier de l’Université de Montréal as previously described (Badr et al. 2008; Cox et al. 2009; Grebely et al. 2012). Subjects who were tested positive for hepatitis B virus or HIV were excluded from the study to prevent confounding of the association between IL28B genotype and HCV clearance. Additional patients undergoing standard-of-care pegylated IFN-α and ribavirin combination therapy were recruited from the Hepatology Department of St-Luc Hospital, Montréal, QC, Canada. This study was approved by the Institutional Ethics Committee (SL05.014, SL05.025) and conducted according to the Declaration of Helsinki. All participants signed an informed consent upon enrolment and, for those included in this study, gave a specific consent for genetic testing. A total of 17 eligible participants (<10 %) refused genetic testing; these were 74.5 % males, 94.1 % Caucasian, and with a mean age of 33.6 years.
Blood sampling and testing
Immediately, 100–200 μl of EDTA blood samples was flash frozen and used for later testing on whole blood. Alternatively, PBMCs were isolated on a Ficoll gradient and cryopreserved. PBMCs were either used directly to extract genomic DNA or to generate EBV-transformed B lymphocyte cell lines (BLCL) as previously described (Tosato and Cohen 2007) as an alternative source of genomic DNA for genotyping.
Genomic DNA isolation
Genomic DNA was extracted from 200 μl of whole blood or 5 × 106 PBMCs or BLCL using a Blood DNA Extraction Kit (Feldan, Quebec City, QC, Canada) following the manufacturer’s protocol.
PCR amplification and IL28B genotyping by sequencing
Isolated genomic DNA was PCR-amplified with standard Taq polymerase (Life Technologies, Carlsbad, CA, USA) using appropriate oligonucleotide primer pairs in a total volume of 50 μl per reaction. PCR conditions were as follows: initial denaturation cycle at 94 °C for 2 min, 40 amplification cycles of 94 °C for 30 s, 55.5 °C for 1 min, and 72 °C for 3 min. A final extension step at 72 °C for 7 min was applied. The PCR-amplified fragments were sequenced by the Sanger Sequencing Service using Applied Biosystem’s 3730xl DNA Analyzer technology at the McGill University and Génome Québec Innovation Centre, Montréal, QC, Canada. The sequencing chromatogram was read to obtain the genotype and to discriminate between homozygotes and heterozygotes.
IL28B screening by PCR
The isolated genomic DNA was PCR-amplified with standard Taq polymerase (Life Technologies, Carlsbad, CA, USA) using appropriate oligonucleotide primer pairs in a total of 25-μl reaction volume. PCR conditions in thermocycler (Biometra, Goettingen, Germany) were as follows: initial denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 65 °C for 15 s, and 72 °C for 1 min for rs12979860 or 30 cycles of 94 °C for 30 s, 54.5 °C for 15 s, and 72 °C for 1 min for rs8099917. A final extension step at 72 °C for 7 min was applied in each case. Appropriate volumes of the PCR reactions were resolved on a 2 % agarose gel and the genotype was determined by the presence of amplified bands of interest.
Statistical analysis
Two measures of linkage disequilibrium (r and |d′|) between rs12979860 and rs8099917 were calculated using the PROC ALLELE procedure implemented in SAS software (SAS Institute, Cary, NC, USA). Haplotype frequencies were estimated while accounting for Hardy–Weinberg equilibrium using the EM algorithm and the PROC HAPLOTYPE procedure implemented in SAS.
Results
Development of a rapid PCR-based screening for IL28B SNPs
We developed a rapid PCR-based test for screening of IL28B SNPs rs12979860 and rs8099917. Primers for IL28B SNP screening were designed from consensus human sequences (NCBI contig NT_011109.16) using the Primer-BLAST tool from the NCBI (Rozen and Skaletsky 2000) and the appropriate SNP mutations. The reverse test primers were designed so that the last nucleotide is the targeted SNP and the primer would therefore anneal or not depending on the patient’s or sample genotype. Primer sequences are listed in Table 1. Genomic DNA was extracted from fresh or frozen whole blood, PBMCs, or BLCLs. Samples were then amplified by PCR with primers specific for each SNP and then resolved on an agarose gel to determine the genotype of each individual. Representative PCR results are demonstrated in Fig. 1. Similar results were obtained irrespective of the source of genomic DNA.
Sample results of the new rapid PCR-based technique. a Sample results for screening of rs12979860 showing a mock PCR, a heterozygote genotype (C/T), a homozygote nonresponder (T/T), and a homozygote responder (C/C). b Sample results for screening of rs8099917 showing a mock PCR, a homozygote nonresponder (G/G), a heterozygote genotype (G/T), a homozygote responder (T/T), and another heterozygote genotype (G/T)
To validate our PCR-based new screening technique, we first sequenced 20 selected participants for each SNP using conventional sequencing protocols, and the results were concordant in 19/20 (95 %) of the tested samples for rs12979860 and 20/20 (100 %) for rs8099917. To further validate the results, 72 participants were retested by PCR–restriction fragment length polymorphism (PCR-RFLP) as previously described (Suppiah et al. 2009); 67 out of 72 results were identical for rs12979860 (93 % concordance) and 71 out of 72 results were identical for rs8099917 (98 % concordance). Taking sequencing as the gold standard, the validity of our test is similar to some of the other rapid tests that were developed recently (Nakamoto et al. 2011). The samples that were discordant between our test and the PCR-RFLP test were retyped by sequencing. These samples were all typed as CC and were typed as CT by our test suggesting that the test might be less powerful in some heterozygote patients.
Distribution of IL28B SNPS in the French Canadian population
Next, we investigated allelic distribution of IL28B SNPs in the Montreal HEPCO cohort of IDUs at high risk of HCV infection. Participants were classified into independent but slightly overlapping populations based on their status at time of recruitment into the cohort:
-
(A)
Base population (n = 183) consists of HCV-naïve individuals at high risk of infection identified as HCV RNA negative and anti-HCV antibody negative at recruitment with one or more risk factor for HCV infection. Assuming that IL28B genotype had no effect on recruitment and selection into this population, it was used as a base population to determine the genetic makeup of the Montreal HEPCO IDU which is representative of the French Canadian IDU population in Montreal.
In order to examine the overall prevalence of the IL28B SNPs in individuals who have cleared HCV either spontaneously or following treatment, two additional populations were formed:
-
(B)
Spontaneous resolvers (n = 67) consists of individuals who resolved spontaneously a previous HCV infection, identified as HCV RNA negative and anti-HCV antibody positive with no prior history of treatment. This included 12 individuals from population (A) who became acutely infected with HCV and spontaneously cleared the virus after joining the cohort as well as 55 other long-term spontaneous resolvers who were already HCV RNA negative and anti-HCV antibody positive at the time of recruitment.
-
(C)
Sustained virological responders (SVRs) (n = 56) consist of individuals who have cleared a previous HCV infection following treatment. This population was identified as HCV RNA negative and anti-HCV antibody positive with documented IFN therapy. This included 12 individuals from population (A) who became persistently infected and underwent treatment following recruitment into the cohort as well as 44 other long-term SVRs who had already achieved sustained viral response at the time of recruitment.
Demographics and patient characteristics are summarized in Table 2. All populations were predominantly Caucasian, male, and with a comparable mean age at recruitment.
Analysis of the overall prevalence of the different IL28B alleles and genotypes in the base population (A) is presented in Table 3. We observed a very high and comparable prevalence of the responder C allele of the rs12979860 SNP (90.2 %) and of the T allele of the rs8099917 (91.3 %). The nonresponder C/T or T/T genotypes, hereafter referred to as */T or nonresponder genotype of rs12979860, were more prevalent than the G/T or G/G genotypes, hereafter referred to as */G or nonresponder genotype of rs8099917 (48.6 vs 36.6 %). When compared to previous studies in similar cohorts, we observed high disparity in the frequency of the nonresponder genotype of the two SNPs resulting in a relatively high proportion of individuals (30.6 %) who do not have the same genotypic status for both SNPs (i.e., rs12979860 is C/T (heterozygote) while rs8099917 is T/T (homozygote responder)) (Table 4 and Fig. 2). The estimated haplotypes, accounting for Hardy–Weinberg equilibrium of each marker in the estimation, showed that the C-T haplotype was the most frequent (61.1 %), followed by T-T (16.3 %), T-G (13.0 %), and C-G (9.7 %). As a result, we observed partial linkage disequilibrium between the two SNPs with |d′| = 0.68 and r = 0.59. This differs greatly from studies done in other population where |d′| is often equal to or very close to 1, including studies done exclusively in Caucasian populations (Lindh et al. 2011; Rauch et al. 2010).
Prevalence of IL28B genotypes in the French Canadian population compared to the US and Australian populations. Left: Prevalence of SNPs rs12979860 and rs8099917 in our study's base population (A). Overlaying shows portions of the population that have different allelic status for rs8099917 and rs12979860. Right: Prevalence of these SNPs in the populations studied in the original GWAS articles of IL28B polymorphism and HCV showing higher prevalence of responder genotype in the French Canadian population
Favorable IL28B SNPs are more prevalent in HCV resolvers
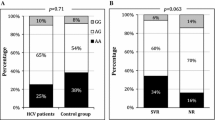
While we observed a higher prevalence of the two responder SNPs as compared to other cohorts, the cumulative incidence of spontaneous viral clearance in this population (18 patients out of 89 of population (A) who were followed during acute HCV infection, 20.2 %) was similar to what was observed in other cohorts (Micallef et al. 2006). Nevertheless, analysis of the overall prevalence of these two SNPs in all spontaneous resolvers (population (B) consisting of 67 spontaneous resolvers) demonstrated a higher prevalence of the rs12979860 responder genotype (58.2 vs 51.4 %) and of the rs8099917 responder genotype (76.1 vs 63.4 %) compared to base population (A). Similarly, analysis of the overall prevalence of the responder SNPs in all sustained viral response patients (population (C) SVRs, consisting of 56 SVRs) demonstrated higher prevalence of the responder genotype C/C of rs12979860 in this subpopulation with (62.5 vs 51.4 %) than in the base population (A); this difference was not as strong in rs8099917 (66.1 vs 63.4 %) (Table 5).
Discussion
Here, we described a rapid PCR-based genetic testing tool for screening of IL28B SNPs rs12979860 and rs8099917 that can be applied to various sample types. While other techniques such as PCR-RFLP, RT-PCR, and direct sequencing exist, they are usually more time consuming and are always more expensive than a simple PCR. This test therefore offers an inexpensive alternative to currently used tests while being as accurate as proven by the comparison with direct sequencing. It would be particularly useful in a research setting or in a cost-constrained setting, as it can be done on a minimal volume of either fresh or frozen whole blood or a low number of patient PBMCs.
The Montreal IDU population is unique compared to previously studied populations in being a predominantly French Canadian population (90.2 % of base population answered White/Caucasian ethnicity in the recruitment questionnaire). The more than six million French Canadians in the province of Quebec are descendants of less than 10,000 French settlers of New France in the seventeenth and eighteenth centuries (Scriver 2001). Genetic studies have identified a strong founder effect in this population (Roy-Gagnon et al. 2011; Scriver 2001). This may explain the higher percentage of responder genotype than in other cohorts (Fig. 2) and the unique disparity in the two responder genotypes resulting in lower linkage disequilibrium than what was observed in other cohorts.
Because of the small numbers in our different subgroups, we could not accurately assess the role of the two IL28B SNPs in spontaneous or therapeutic viral clearance. Other covariates, such as gender or HCV genotype, could not be assessed as the cohort was predominantly male and many patients could not be genotyped because of limited sample availability. Nevertheless, we observed that the spontaneous viral clearance population was enriched in individuals with the responder genotype. We also observed that, in our specific population, rs12979860 is more variable (nonresponder genotype present in 48.6 % of population vs 36.6 % for rs8099917). As a consequence, rs12979860 might be a better predictor of clearance following treatment in this population. This warrants an investigation in a larger cohort where other covariates including other genetic factors or SNPs could be examined.
Despite the higher prevalence of the two responder SNPs as compared to other cohorts (Fig. 2), this was not associated with a better rate of spontaneous viral clearance in acute HCV-infected individuals of population (A), as compared to results from other cohorts (20.2 vs 26 %, respectively) (Micallef et al. 2006). This could be due to several factors. First is the lower linkage disequilibrium between the two responder SNPs or other host genetic factors like the recently identified IFNL4 gene located upstream of the IL28B gene (Prokunina-Olsson et al. 2013). It is also possible that the rate of clearance is influenced by the fact that all our participants are IDUs with potential lifestyle and socioeconomic problems. Other potential factors could be viral load and viral evolution within that specific population (Liu et al. 2012). Immune factors might also be involved as demonstrated by the use of baseline IP-10 serum levels (Beinhardt et al. 2012). It also has been suggested that IL28B polymorphism may influence outcome of acute HCV indirectly by modulating NK cell activity (Dring et al. 2011; Suppiah et al. 2011). Therefore, host genetics is an important predictor of outcome of acute HCV infection, but its predictive value would be greatly enhanced by considering other factors such as the host immune response and viral variation, especially in smaller study populations.
References
Alter MJ (2007) Epidemiology of hepatitis C virus infection. World J Gastroenterol 13:2436–2441
Badr G, Bedard N, Abdel-Hakeem MS, Trautmann L, Willems B, Villeneuve JP, Haddad EK, Sekaly RP, Bruneau J, Shoukry NH (2008) Early interferon therapy for hepatitis C virus infection rescues polyfunctional, long-lived CD8+ memory T cells. J Virol 82:10017–10031
Barreiro P, Vispo E, Poveda E, Fernandez-Montero JV, Soriano V (2012) Hepatitis C therapy—highlights from EASL 2012. Clin Infect Dis 56(4):560–566
Beinhardt S, Aberle JH, Strasser M, Dulic-Lakovic E, Maieron A, Kreil A, Rutter K, Staettermayer AF, Datz C, Scherzer TM, Strassl R, Bischof M, Stauber R, Bodlaj G, Laferl H, Holzmann H, Steindl-Munda P, Ferenci P, Hofer H (2012) Serum level of IP-10 increases predictive value of IL28B polymorphisms for spontaneous clearance of acute HCV infection. Gastroenterology 142(78–85):e2
Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID (2005) Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128:1437
Cox AL, Page K, Bruneau J, Shoukry NH, Lauer GM, Kim AY, Rosen HR, Radziewicz H, Grakoui A, Fierer DS, Branch AD, Kaplan DE, Chang KM (2009) Rare birds in North America: acute hepatitis C cohorts. Gastroenterology 136:26–31
Darling JM, Aerssens J, Fanning G, McHutchison JG, Goldstein DB, Thompson AJ, Shianna KV, Afdhal NH, Hudson ML, Howell CD, Talloen W, Bollekens J, De Wit M, Scholliers A, Fried MW (2011) Quantitation of pretreatment serum interferon-gamma-inducible protein-10 improves the predictive value of an IL28B gene polymorphism for hepatitis C treatment response. Hepatology 53:14–22
Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R, O'Farrelly C, Gardiner CM (2011) Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc Natl Acad Sci U S A 108:5736–5741
Dustin LB, Rice CM (2007) Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol 25:71–99
Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401
Grebely J, Morris MD, Rice TM, Bruneau J, Cox AL, Kim AY, McGovern BH, Shoukry NH, Lauer G, Maher L, Lloyd AR, Hellard M, Prins M, Dore GJ, Page K, on behalf of the InC3 Study Group (2012) Cohort profile: the International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) study. Int J Epidemiol. doi:10.1093/ije/dys167
Heathcote EJ (2004) Prevention of hepatitis C virus-related hepatocellular carcinoma. Gastroenterology 127:S294–S302
Holmes JA, Desmond PV, Thompson AJ (2012) Does IL28B genotyping still have a role in the era of direct-acting antiviral therapy for chronic hepatitis C infection? J Viral Hepat 19:677–684
Hoofnagle JH (2002) Course and outcome of hepatitis C. Hepatology 36:S21–S29
Lindh M, Lagging M, Arnholm B, Eilard A, Nilsson S, Norkrans G, Soderholm J, Wahlberg T, Wejstal R, Westin J, Hellstrand K (2011) IL28B polymorphisms determine early viral kinetics and treatment outcome in patients receiving peginterferon/ribavirin for chronic hepatitis C genotype 1. J Viral Hepat 18:e325–e331
Liu L, Fisher BE, Thomas DL, Cox AL, Ray SC (2012) Spontaneous clearance of primary acute hepatitis C virus infection correlated with high initial viral RNA level and rapid HVR1 evolution. Hepatology 55:1684–1691
Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D, Carretta V, Minerva N, Goldstein DB, McHutchison JG (2010) An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology 139:821–827, 827 e1
Micallef JM, Kaldor JM, Dore GJ (2006) Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat 13:34–41
Nakamoto S, Kanda T, Imazeki F, Wu S, Arai M, Fujiwara K, Yokosuka O (2011) Simple assay based on restriction fragment length polymorphism associated with IL28B in chronic hepatitis C patients. Scand J Gastroenterol 46:955–961
Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR (2013) A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45:164–171
Rallon NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, Shianna KV, Vispo E, Thompson A, McHutchison J, Soriano V (2010) Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS 24:F23–F29
Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Gunthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Mullhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY (2010) Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138:1338–1345, 1345 e1-7
Roy-Gagnon MH, Moreau C, Bherer C, St-Onge P, Sinnett D, Laprise C, Vezina H, Labuda D (2011) Genomic and genealogical investigation of the French Canadian founder population structure. Hum Genet 129:521–531
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Scriver CR (2001) Human genetics: lessons from Quebec populations. Annu Rev Genomics Hum Genet 2:69–101
Shoukry NH, Cawthon AG, Walker CM (2004) Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol 58:391–424
Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J (2009) IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41:1100–1104
Suppiah V, Gaudieri S, Armstrong NJ, O'Connor KS, Berg T, Weltman M, Abate ML, Spengler U, Bassendine M, Dore GJ, Irving WL, Powell E, Hellard M, Riordan S, Matthews G, Sheridan D, Nattermann J, Smedile A, Muller T, Hammond E, Dunn D, Negro F, Bochud PY, Mallal S, Ahlenstiel G, Stewart GJ, George J, Booth DR, International Hepatitis CGC (2011) IL28B, HLA-C, and KIR variants additively predict response to therapy in chronic hepatitis C virus infection in a European Cohort: a cross-sectional study. PLoS Med 8:e1001092
Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M (2009) Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801
Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R, Poordad F, Lawitz EJ, McCone J, Shiffman ML, Galler GW, Lee WM, Reindollar R, King JW, Kwo PY, Ghalib RH, Freilich B, Nyberg LM, Zeuzem S, Poynard T, Vock DM, Pieper KS, Patel K, Tillmann HL, Noviello S, Koury K, Pedicone LD, Brass CA, Albrecht JK, Goldstein DB, McHutchison JG (2010) Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology 139(120–9):e18
Tosato G, Cohen JI (2007) Generation of Epstein–Barr virus (EBV)-immortalized B cell lines. Curr Protoc Immunol Chapter 7:Unit 7.22
Walker CM (2010) Adaptive immunity to the hepatitis C virus. Adv Virus Res 78:43–86
Acknowledgments
We thank all our blood donors as well as the HEPCO team for their help with participant recruitment and follow-up. We thank Marie-Pierre Sylvestre and Youssef Idaghdour for the assistance in biostatistical analysis. This study was supported by grants from the Canadian Institutes for Health Research (CIHR) (MOP-106468), the Canadian Liver Foundation, and the Fonds de recherche du Québec-Santé (FRQS) AIDS and Infectious Disease Network (Réseau FRQS SIDA/MI). T. Fabre is a recipient of a graduate fellowship from Fondation du Dr Gabriel Marquis and Faculté des études supérieures et postdoctorales, Université de Montréal. P. Willems received a summer student fellowship from the National CIHR Research Training Program in Hepatitis C. J. George is supported by the Storr Bequest to the Sydney Medical Foundation and grants from the NHMRC. N. H. Shoukry and J. Bruneau are the recipients of Chercheur Boursier salary awards from FRQS.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s00251-015-0831-8.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Gélinas, JF., Fabre, T., Willems, P. et al. IL28B SNP screening and distribution in the French Canadian population using a rapid PCR-based test. Immunogenetics 65, 397–403 (2013). https://doi.org/10.1007/s00251-013-0688-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-013-0688-7