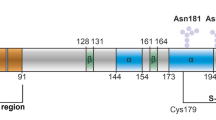
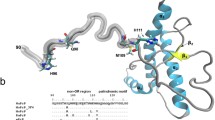
Abstract
In this work we present and analyse XAS measurements carried out on various portions of Prion-protein tetra-octa-repeat peptides in complexes with Cu(II) ions, both in the presence and in the absence of Zn(II). Because of the ability of the XAS technique to provide detailed local structural information, we are able to demonstrate that Zn acts by directly interacting with the peptide, in this way competing with Cu for binding with histidine. This finding suggests that metal binding competition can be important in the more general context of metal homeostasis.










Similar content being viewed by others
Notes
For a more detailed description of how the χ(k) EXAFS signal is extracted from XAS data see the Appendix in Minicozzi et al. (2008).
The normalization is performed in a standard way using the ATHENA software (Ravel 2008), i.e. fitting the pre-edge region with a straight line and subtracting it from the whole spectrum (Ravel and Newville 2005). The post-edge region data are then fitted with a polynomial. Finally, the jump between the pre-edge and the post-edge fits at the edge energy is calculated and the spectrum is divided by the height of the jump. We recall the standard formulae \( {\chi (k) = \frac{{\mu (E) - \mu_{0} (E)}}{{\mu_{0} (E)}}} \) and \( {\hbar k = \sqrt {2m(E - E_{0} )} } \) where μ 0(E) is the single atom absorption coefficient and E 0 is the edge energy.
For completeness we provide in the supplementary material a quantitative analysis of the EXAFS spectra of samples S3 and S3_Zn data at the Cu K-edge. The analysis confirms that the structure of the Cu2+ binding site is not affected by the presence of Zn2+ and the fitted site geometry is highly consistent with the available crystallographic information (Chattopadhyay et al. 2005).
It should be said that here we have simplified the analysis by assuming that component β corresponds to a situation in which all four His units of each tetra-octa-repeat are simultaneously involved in binding the metal. For the sake of completeness, the same analysis has also been performed assuming that Cu2+ in component β is bound to three His units. The results obtained under this assumption do not differ from those presented here in the text and can be found in the supplementary material.
Here we make the further hypothesis that the reason why Zn2+ can be found free in solution it is that there are no more available His units.
The quality factor R is computed by use of the formula: \( {\text{R}} = \sum\nolimits_{{{\text{i}} = 1}}^{\text{P}} {\frac{ 1}{{{\text{w}}_{\text{i}} }}} \left| {{{\upchi}}^{ \exp } (k_{\text{i}} )- {{\upchi}}^{\text{fit}} (k_{\text{i}} )} \right| \) where P is the number of experimental points and \( {\text{w}}_{\text{i}} = \frac{1}{{k_{i}^{n} }}\sum\nolimits_{j = 1}^{P} {k_{j}^{n} |{{\upchi}}^{\exp } (k_{j} )| \, } \) in which n is an integer which is normally taken to lie between 0 and 3 (here we took n = 3).
In all the fits only the distance and relative position of the nearest neighbouring atoms in the His-bound Zn2+ environment are taken as free variables.
Incidentally, we have also tried to leave the Zn2+ fraction in buffer as a free variable in the fit to the S1_Zn XAS spectra (data not shown). Values in good agreement with the numbers in the second column of Table 2 are obtained when the structural models M1 and/or M2 are assumed. Completely unrealistic values are instead selected by the fit with models M3 and M4.
In the supplementary material the same plot as in Fig. 9 is drawn by also taking into account the errors in the 〈r in〉 determination. In this way both M1 and M2 models yield numbers for 〈r in〉 that fall within the range of the acceptable BVS values as defined in the text.
References
Aronoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, Antholine WE, Ball HL, Cohen FE, Prusiner SB, Millhauser GL (2000) Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry 39:13760–13771
Binsted N (1998) EXCURV98. CCLRC Daresbury Laboratory, Warrington, Cheshire, UK
Binsted N, Strange RW, Hasnain SS (1992) Constrained and restrained refinement in EXAFS data analysis with curved wave theory. Biochemistry 31:12117–12125
Brown ID (2010) Private communication (see also www.mrl.ucsb.edu/~seshadri/2009_218/bvparm2006.pdf)
Brown ID, Altermatt D (1985) Bond-valence parameters obtained from a systematic analysis of the inorganic crystal structure database. Acta Cryst B4:244–247
Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, Antholine WE, Olmstead MM, Vrielink A, Gerfen GJ, Peisach J, Scott WG, Millhauser GL (2002) Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry 41:3991–4001
Burns CS, Aronoff-Spencer E, Legname G, Prusiner SB, Antholine WE, Gerfen GJ, Peisach G, Millhauser GL (2003) Copper coordination in the full-length, recombinant prion protein. Biochemistry 42:6794–6803
Cereghetti GM, Schweiger A, Glockshuber R, Van Doorslaer S (2001) Electron paramagnetic resonance evidence for binding of Cu2+ to the C-terminal domain of the murine prion protein. Biophys J 81:516–525
Chattopadhyay M, Walter ED, Newell DJ, Jackson PJ, Aronoff-Spencer E, Peisach J, Gerfen GJ, Bennett B, Antholine WE, Millhauser GL (2005) The octarepeat domain of the prion protein binds Cu(II) with three distinct coordination modes at pH 7.4. J Am Chem Soc 127(36):12647–12656
D’Angelo P, Benfatto M, Della Longa S, Pavel NV (2002a) Combined XANES and EXAFS analysis of Co2+, Ni2+, and Zn2+ aqueous solutions. Phys Rev B 66:064209
D’Angelo P, Barone V, Chillemi G, Sanna N, Meyer-Klaucke W, Pavel NV (2002b) Hydrogen and higher shell contributions in Zn2+, Ni2+, and Co2+ aqueous solutions: an X-ray absorption fine structure and molecular dynamics study. J Am Chem Soc 124:1958–1967
Dominikus AL, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, von Schroetter C, Fiorito F, Herrmann T, Güntert P, Wüthrich K (2005) Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci USA 102:640–645
Eghiaian F, Grosclaude J, Lesceu S, Debey P, Doublet B, Tréguer E, Rezaei H, Knossow M (2004) Insight into the PrPC→PrP(SC) conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc Natl Acad Sci USA 101:10254–10259
Ferreira GC, Franco R, Mangravita A, George GN (2002) Unraveling the substrate-metal binding site of ferrochelatase: an X-ray absorption spectroscopic study. Biochemistry 41:4809–4818
Fonda L (1992) Multiple-scattering theory of X-ray absorption: a review. J Phys Condens Matter 4:8269–8302
Gasset M, Baldwin MA, Fletterick RJ, Prusiner SB (1993) Perturbation of the secondary structure of the scrapie prion protein under conditions that alter infectivity. Proc Natl Acad Sci USA 90:1–5
Guerrieri F, Minicozzi V, Morante S, Rossi GC, Furlan S, La Penna G (2009) Modeling the interplay of glycine protonation and multiple histidine binding of copper in the prion protein octarepeat sub-domains. J Biol Inorg Chem 14:361–374
Hasnain SS, Murphy LM, Strange RW, Grossmann JG, Clarke AR, Jackson GD, Collinge J (2001) XAFS Study of the High-affinity Copper-binding Site of Human PrP91–231 and its low-resolution structure in solution. J Mol Biol 311:467–473
Hornshaw MP, McDermott JR, Candy JM, Lakey JH (1995) Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem Biophys Res Commun 214:993–999
Minicozzi V, Stellato F, Comai M, Dalla Serra M, Potrich C, Meyer-Klaucke W, Morante S (2008) Identifying the minimal Cu and Zn binding site sequence in amyloid beta peptides. J Biol Chem 283:10784–10792
Morante S (2008) Metal ions and protein aggregation: the case of prion protein and β-amyloids (invited review). In: Bulone D, San Biagio PL (eds) Biophysical inquiry into protein aggregation and amyloid diseases, research signpost edition. Transworld Research Network, Kerala, pp 53–110
Morante S, González-Iglesias R, Potrich C, Meneghini C, Meyer-Klaucke W, Menestrina G, Gasset M (2004) Inter- and intra-octarepeat Cu(II) site geometries in the prion protein. Implication in Cu(II) binding cooperativity and Cu(II)-mediated assemblies. J Biol Chem 279:11753–11759
Nunziante M, Gilch S, Schatzl HM (2003) Essential rôle of the prion protein N terminus in subcellular trafficking and half-life of cellular prion protein. J Biol Chem 278:3726–3734
Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, Prusiner SB (1993) Conversion of α-Helices β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA 90:10962–10966
Proux O, Biquard X, Lahera E, Menthonnex J–J, Prat A, Ulrich O, Soldo Y, Trévisson P, Kapoujvan G, Perroux G, Taunier P, Grand D, Jeantet P, Deleglise M, Roux J-P, Hazemann J-L (2005) FAME: A new beamline for X-ray absorption investigations of very-diluted systems of environmental, material and biological interests. Phys Scripta 115:970–973
Proux O, Nassif V, Prat A, Ulrich O, Lahera E, Biquard X, Menthonnex J–J, Hazemann J-L (2006) Feedback system of a liquid nitrogen cooled double-crystal monochromator: design and performances. J Synchrotron Radiat 13:59–68
Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95:13363–13383
Qin K, Yang Y, Mastrangelo P, Westaway D (2002) Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J Biol Chem 277:1981–1990
Rachidi W, Mange A, Senator A, Guiraud P, Riondel J, Benboubetra M, Favier A, Lehmann S (2003) Prion infection impairs copper binding of cultured cells. J Biol Chem 278:14595–14598
Ravel B (2008) ATHENA user’s guide. http://cars9.uchicago.edu/ravel/software/exafs/
Ravel B, Newville M (2005) ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J Synchrotron Rad 12:537–541
Stockel J, Safar J, Wallace AC, Cohen FE, Prusiner SB (1998) Prion protein selectively binds copper(II) ions. Biochemistry 37:7185–7193
Strange RW, Alagna L, Durham P, Hasnain SS (1990) An understanding of the x-ray absorption near-edge structure of copper(II) imidazole complexes. J Am Chem Soc 112:4265–4268
Thorp HH (1992) Bond valence sum analysis of metal-ligand bond lengths in metalloenzymes and model complexes. Inorg Chem 31:1585–1588
Walter ED, Stevens DJ, Visconte MP, Millhauser GL (2007) The prion protein is a combined zinc and copper binding protein: Zn2+ alters the distribution of Cu2+ coordination modes. J Am Chem Soc 129:15440–15441
Wells MA, Jelinska C, Hosszu LLP, Craven CJ, Clarke AR, Collinge J, Waltho JP, Jackson GS (2006) Multiple forms of copper (II) co-ordination occur throughout the disordered N-terminal region of the prion protein at pH 7.4. Biochem J 400:501–510
Whittal RM, Ball HL, Cohen FE, Burlingame AL, Prusiner SB, Baldwin MA (2000) Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci 9:332–343
Acknowledgments
We are very grateful to G.C. Rossi for useful discussions and for reading the manuscript. Partial financial support from PRIN08 is acknowledged. We thank the anonymous referees for their useful suggestions.
Author information
Authors and Affiliations
Corresponding author
Additional information
F. Stellato, A. Spevacek contributed equally to the work.
Special Issue: SIBPA Meeting 2011.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stellato, F., Spevacek, A., Proux, O. et al. Zinc modulates copper coordination mode in prion protein octa-repeat subdomains. Eur Biophys J 40, 1259–1270 (2011). https://doi.org/10.1007/s00249-011-0713-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-011-0713-4