Abstract
Here we review the role played by transient interactions between multi-functional proteins and their RNA targets in the regulation of mRNA metabolism, and we describe the important function of NMR spectroscopy in the study of these systems. We place emphasis on a general approach for the study of different features of modular multi-domain recognition that uses well-established NMR techniques and that has provided important advances in the general understanding of post-transcriptional regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Transient interactions in post-transcriptional regulation
In eukaryotes, the majority of genes are regulated both at the transcriptional and post-transcriptional level. Post-transcriptional control of gene expression is essential for cell differentiation and cell function and—in complex organisms—is provided by a highly sophisticated regulatory network where the steps of mRNA metabolism (mRNA capping, poly-adenylation, splicing, degradation, etc.) and mRNA localisation are coordinated to modulate local protein concentration.
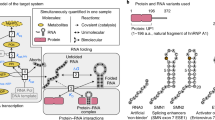
The coordination between the different processes relies on the assembly of large mRNA-protein complexes—known as ribonucleoprotein particles (RNPs)—that migrate between the sites of processing, storage and translation. RNP particles have been grouped in classes reflecting their protein composition and their association with specific cellular states, and a general regulatory model has been proposed where functionally related genes are co-regulated based on the association of the corresponding mRNAs within the same RNP particles (Keene and Tenenbaum 2002; Mansfield and Keene 2009). In this so-called ‘ribonome’ model, RNP particles represent the functional equivalent of bacterial operons (Fig. 1). The composition of each RNP particle varies depending on its functional status, and the multi-functional multi-domain protein components can associate with different RNA molecules in a timely and localised fashion. These protein-RNA interactions control the efficiency of mRNA synthesis, processing, nuclear export and degradation as well as the mRNA’s translation rate and cellular localisation (Keene and Tenenbaum 2002; Mansfield and Keene 2009).
The ribonome. In eukaryotic cells, ensembles of mRNA molecules are co-regulated within RNPs. RNPs are dynamic protein-RNA particles, where different proteins engage in the regulation of the metabolism and transport of mRNA molecules (left). Recently, small non-coding RNAs (snRNAs)—that are themselves regulated by dedicated pathways (right)—have been shown to play an important role in mRNA regulation
Reversibility is a key feature of the protein-RNA interactions described above and is achieved by the use of a modular approach where different domains of the same protein make contact with the RNA target or where different interacting proteins bind synergistically to the same RNA target. Modular interactions—where the affinity of each domain for the RNA is low—are easier to reverse than a single high affinity one and lend themselves to being regulated by small changes in the concentration of the protein(s) involved (Lunde et al. 2007). Interestingly, the individual binding modules can play different roles in the recognition of different targets. For example, in the KSRP system the KH3 domain plays a dominant role in the recognition of a G-rich target linked to the regulation of miRNA biogenesis, while its affinity for the AU-rich elements that mediate recruitment of the mRNA degradation machinery to target mRNAs is closer to the one of the other domains (García-Mayoral et al. 2008). Therefore, understanding modular protein-RNA interactions is a challenging process.
A clear link between the ensemble of in vivo targets of multi-domain multifunctional regulators identified by high-throughput screenings—such as CLIP (Ule et al. 2003)—and the in vitro data on binding affinity and specificity is, with a few exceptions (Buckanovich and Darnell 1997; Licatalosi et al. 2008; Llorian et al. 2010; Reid et al. 2009; Yeo et al. 2009), yet to be established. Our current understanding of the interactions indicate that a structural and biophysical description of the RNA- and protein-binding modules is not sufficient for the prediction of target specificity of most multi-domain proteins in the cell, and a full account of their target preference and of the multiple interactions that tune the basic RNA binding capability of the domains is necessary.
Structure and dynamics of RNA-binding domains
RNA-binding domains are the defining feature of the multi-functional RNA-binding proteins regulating mRNA metabolism and are often present in multiple copies within the same protein (Lunde et al. 2007). Domains from a small number of RNA-binding folds—RRM, KH, dsRBD, S1, ZnF, etc.—account for the RNA-binding capability of most known RNA-binding proteins. These domains are small—between 50 and 100 amino acids—and the structure of several members of each family has been characterised by X-ray crystallography and solution NMR spectroscopy (Bycroft et al. 1997; Clery et al. 2008; Doyle and Jantsch 2003; Hall 2005; Valverde et al. 2008). This structural analysis has shown that in many of the domains a common fold is flanked by additional structural elements that provide an important input to RNA recognition (Allain et al. 1996; Conte et al. 2000; De Guzman et al. 1998; Garcia-Mayoral et al. 2007; Leulliot et al. 2004; Liu et al. 2001; Oberstrass et al. 2005; Pancevac et al. 2010).
In many well-studied examples, RNA-binding domains recognise specific RNA structures or relatively long sequences with Kd in the nanomolar range. Most of the high-resolution structural information on RNA-binding domains in complex with the RNA targets has been obtained on these higher affinity-higher specificity complexes—which are more easily tractable by both NMR and X-ray crystallography. Information on binding affinity and specificity in these systems can also be obtained using several techniques, including EMSA, ITC, Biacore, etc. However, the majority of isolated RNA-binding domains recognise their RNA targets with Kds in the micromolar range, and the range of techniques that can be used to analyse these interactions is more limited.
NMR is an effective tool to describe the conformation, stability and motions of the RNA-binding domains above. The domains’ size is well within the range of standard NOE and residual dipolar coupling (RDC)-based techniques for structure determination. It also allows the study of backbone and side chain dynamics in a picoseconds to milliseconds range using well-established techniques. Most studies of the motions taking place in RNA-binding proteins rely on the use of 15N-labelling and of T1, T2 and heteronuclear NOE experiments that report on fast (nanosecond—T1 and heteronuclear NOE) and slow (millisecond—T2) motions in the protein backbone. Motions in the millisecond timescale, which can be related to large movements of secondary structure elements sometimes observed upon RNA binding, are instead monitored using recently introduced relaxation dispersion experiments (Deka et al. 2005; Tollinger et al. 2001).
NMR relaxation experiments can be used to characterise motions related to both high-affinity/high-specificity protein-RNA interactions and to the more transient protein-RNA interactions this review focuses on. The characterisation of the dynamic behaviour of a number of RNA-binding domains has shown that motions take place not only in the protein loops—that often rearrange upon RNA binding—but also in highly dynamic spots that are sometimes present within the core structural elements both in the free and bound protein. Interestingly, the interaction of the domain’s core with the flanking structural elements is often transient, and in crystals these peripheral elements can engage in macromolecular packing that can stabilise one of several conformations (Andrec et al. 2007; Pancevac et al. 2010). NMR is uniquely suited to examine these interactions. Indeed, NMR studies have shown that flanking elements can control the access to the RNA-binding surface in a number of RRM domains (Allain et al. 1996; Pancevac et al. 2010), extend the RNA-binding surface in KH, ZnF and RRM domains (e.g. Conte et al. 2000; De Guzman et al. 1998; Liu et al. 2001; Oberstrass et al. 2005), and stabilise the RNA-binding elements in a conformation optimal for binding in dsRBD and KH domains (Garcia-Mayoral et al. 2007; Leulliot et al. 2004). In some of the best studied cases, such as the one of the U1A in complex with the PIE element of its own 3′UTR, protein-protein and protein-RNA interactions combine to mediate regulation. Here binding of U1A to the symmetrical PIE RNA target releases a carboxi-terminal helix from the RNA-binding surface. This helix acts as a protein dimerization element and, at the same time, as a recruiting element for poly(A)polymerase (Varani et al. 2000). Finally, understanding of the motions within the inter-domain linker is important to rationalise the binding cooperativity between RNA-binding domains and their recognition of different RNA structures.
RNA recognition by RNA-binding domains
The high-resolution structures of a number of domains in complex with their minimal RNA targets have been solved by NMR and X-ray crystallography. These structural data have allowed us to draw some general conclusions on protein-RNA interaction interfaces, i.e. that these interfaces are positively charged, highly hydrated and enriched in aromatic residues, when compared with protein-protein interaction surfaces. They are also enriched in hydrogen bonds between the protein moieties and the RNA phosphate groups, the specific 2’OH group of the sugar and the RNA bases, the latter being very important in defining sequence specificity (Bahadur and Zacharias 2008; Ellis et al. 2007; Treger and Westhof 2001). Protein-RNA interfaces are often dynamic, and the interaction itself results in conformational changes within the interface (Dominguez et al. 2011).
NMR can describe the structure and dynamics of transient protein-RNA complexes and characterise their binding affinity and specificity. NMR has been used to solve the hig-resolution structure of protein-RNA complexes with Kds in the μM range, an affinity range that includes many of the transient interactions between ssRNA-binding domains and short RNA targets (Fig. 2a, Auweter et al. 2006). These structures have been solved using a standard set of NMR experiments and calculation protocols, although specific issues (such as the calibration of NOE-derived inter-molecular restraints) must be considered and ad hoc experimental strategies (for example involving the probing of different RNA constructs and the use of low salt) may be required. Additionally, docking programmes have been introduced in the structure calculation protocol to improve convergence and to allow the use of non-NOE restraints for the determination of lower resolution structures (Martin-Tumasz et al. 2010).
NMR studies of protein-RNA interactions. a The high-resolution structure of the PTB RRM2 (green, cartoon)—5′-CUCUCU-3′ (orange, sticks) complex (PDB: 2ADB). The RNA binds to the canonical RNA-binding surface of the RRM domain that is however extended by an additional, fifth β-strand, C-terminal to the core fold of the protein. b Ribbon representation of CstF-64 RRM domain (PDB: 1P1T). A C-terminal α-helix, which in the free protein is packed against the β-sheet surface covering the RNA-binding site and which unwinds upon RNA binding, is not displayed. The residues of the RNA-binding surface whose backbone dynamics change upon RNA binding are highlighted in blue (Deka et al. 2005). c Measuring affinity by NMR. Superimposed HSQC spectra (in different colours) recorded during titration of KSRP KH3 domain with 5′-AGGGU-3′ RNA. In the inset the shift in the position of a peak is plotted against the RNA/protein ratio (empty circles) to calculate the binding curve (in red). d SIA scores obtained for KSRP KH3. The domain binds preferentially to G-rich sequences. This graphic representation was generated by plotting SIA data with the Weblogo server (http://weblogo.berkeley.edu/logo.cgi). e Complementary methods can be used to identify and validate interaction surfaces to be used in design of functional studies and macromolecular docking
In NMR, the most common strategy to analyse the changes in backbone and side chain motions associated to protein-RNA interactions compares two sets of backbone relaxation experiments recorded on the free and bound protein. For example, relaxation dispersion experiments to evaluate protein dynamics on a ms time scale recorded on the transient protein-RNA complex between the Cstf64 protein and GU-rich elements have shown that flexibility in the interface is maintained upon binding (Fig. 2b), suggesting how different nucleobases can be accommodated along the interface and explaining the partially degenerate target sequences (Deka et al. 2005). In a second very different example, comparative NMR studies of the Quaking and SF1 proteins have shown that the QUA2 helix is already formed in the free protein, but not rigidly positioned with respect to the core of the domain. This indicates that the RNA target interacts transiently with the KH domain and is then locked into position by the interaction with the QUA2 helix (Liu et al. 2001; Maguire et al. 2005). Often, very specific high-affinity interactions are coupled to dynamic areas on the protein, and the RNA is locked in a more stable conformation in the complex (Mittermaier et al. 1999; Shajani et al. 2007), while in many lower affinity less specific complexes, the interaction surface remains dynamic (Deka et al. 2005). A dynamic interface allows the recognition of different bases and has also been proposed to decrease the entropic cost of binding, which would favour reversibility (Deka et al. 2005). However, an increase in overall conformational entropy upon binding has also been observed for a number of interactions, and it has been suggested that this may represent a general way to compensate for the loss of entropy within the binding surface (Ravindranathan et al. 2010). Interestingly, it has also been proposed that the unfavourable entropy of protein-RNA interaction can be offset by coupling this interaction to protein-protein interactions within a multi-component system (Ramos et al. 2002).
NMR can be used as a general biophysical tool to measure accurately the strength of protein RNA interactions in complexes with dissociation constants in the μM range using interaction-dependent changes in chemical shift. In these complexes, the position of peaks in a fast regime of exchange on a chemical shift time scale is directly linked to the molar fraction of bound protein. The change in chemical shift of these resonances can be plotted against the RNA:protein ratio and used to fit a binding isotherm and calculate a dissociation constant for the complex (Fig. 2c). The aromatic rings present in protein-RNA interfaces ensure that large chemical shift changes take place upon interaction and facilitate this analysis. It is important to point out that, thanks to the use of super-cooled probes and of other hardware advances, these NMR experiments can be recorded at concentrations ranging from 10–20 μM to a few mM. This allows a direct measure of the Kd across the micromolar range, which is not easily achievable with other techniques.
Recently NMR has also been used to explore the sequence specificity in protein-RNA interactions. Scaffold independent analysis (SIA) defines the nucleobase preference of a protein-binding domain for each of the positions of the bound sequence (Beuth et al. 2007) (Fig. 2d). The method is unbiased and is based on the comparative analysis of the binding affinity of a set of four RNA oligo pools for each position to be screened. SIA has been shown to accurately identify nucleobase specificity in protein-RNA interactions with submillimolar affinity, an affinity that is too low for other equivalent methods to work effectively, but that is common in RNA-binding domains. SIA has been used to successfully predict the sequence specificity of the second RRM domain of the Prp24 protein and to identify its exact binding site on U6 RNA as well as to dissect the sequence specificity of the four KH domains of the KSRP protein (Beuth et al. 2007; Garcia-Mayoral et al. 2008; Martin-Tumasz et al. 2010).
Importantly, NMR experiments can be used to easily monitor the chemical microenvironment of a large set of nuclei in the so-called chemical shift perturbation (CSP) analysis. Sensitive 1H-15N correlation spectra provide a set of reporters (the backbone amide groups) that are well dispersed in the spectrum and across the protein structure. Changes in the position and line shape of these resonances upon RNA binding define the binding interface and allow monitoring of binding and stability. CSP data can therefore be used to facilitate the design of mutants that disrupt the RNA (or protein) binding capability of domains without perturbing the protein stability and aggregation properties. Such mutants represent an ideal tool to be used in functional studies (Fig. 2e). The use of paramagnetic relaxation enhancement (PRE), where a paramagnetic tag is attached to a specific nucleotide on the RNA and the enhanced relaxation of protein resonances measured, or of cross-saturation where the RNA is specifically irradiated and the saturation transfer detected on the protein resonances at the interface, provides a complementary tool to cross-check the interface mapping obtained by CSP data. If a specific study requires confirmation of the mutant domain’s folding, an RDC-based approach to structural monitoring has been proposed (Kirkpatrick et al. 2009). It is worth mentioning that contact information obtained from CSP, PRE and cross-saturation experiments can be used in the docking of protein and RNA molecules to obtain models of very transient complexes.
Combinatorial protein-RNA interactions
The use of a modular domain-based approach to RNA recognition allows regulatory proteins to establish reversible interactions with the RNA targets within large protein-RNA particles. This modularity is observed in the bimolecular recognition of an RNA molecule by a multi-domain protein (Fig. 3a, b) or when several RNA molecules or proteins are involved. Protein-protein interactions can also bring together different parts of the RNA or different RNAs (Fig. 3a, b). A well-studied example of synergistic recognition of an RNA by several RNA-binding proteins is the recognition of the intronic mRNA branch point by branch point recognition complex (Fig. 3c).
Modular recognition of the RNA. a Two RNA-binding domains can interact with adjacent RNA sequences (top) or distantly located sequences within one RNA molecule (middle) or two different RNA molecules (bottom), creating an array of possible structural combinations. b The different RNA-binding affinities of the four KH domains of KSRP for RNA (best binding sequence) underscore their different role and the potential to create a high-affinity interaction in multiple domains recognition. c Schematic representation of the intronic mRNA branch point recognition by the branch point recognition complex (SF1-U2AF65-U2AF35). SF1, splicing factor 1; U2AF65 and U2AF35, U2 snRNP auxiliary factor subunits (65 kDa and 35 kDa); BPS, branchpoint sequence; Py-tract, polypyrimidine tract; AG, 3′ splice site; RRM, RNA recognition motif; UHM, U2AF homology motif; ULM, UHM ligand motif
A mechanistic model that explains recognition between the components of the protein-RNA regulatory machinery must consider a number of simultaneous interactions when describing structural features, energetics and motions of the binding. Building such a model requires a multi-disciplinary approach. NMR’s high information content and broad range of observables offer the possibility to deconstruct the regulatory complexes to simpler two-component systems, while its capability to directly report on a broad range of macromolecular motions is important to understand structural rearrangements, entropic changes and binding models. The basic strategy for the analysis of multiple interactions in high molecular weight complexes is a comparative one, where the information obtained on the single interactions is used to assess the changes that may take place in the larger system. A key advantage of NMR is that the large number of the spatially well-distributed reporters in 1H 15N and 1H 13C fingerprint spectra allow directly analysing simultaneous macromolecular interactions by tracking the reporters from the different interaction surfaces. This analysis includes not only the structural validation of the interaction surfaces itself (Fig. 4a), but also an analysis of backbone dynamics and binding affinity. For example, it is possible to establish if a change in dynamics takes place outside the interaction surface, if the stability of a domain changes upon interaction or if the strength of the interactions between two molecules changes in a three component systems. This information can be used to build a molecular picture of recognition. Such a strategy was used to show that PTB RRM2 independently interacts with its target RNA 5′-CUUCUCUCU-3′ and the regulatory protein Raver1 (Rideau et al. 2006). Also, a recent study showed that in the protein-protein-DNA FBP-FIR-FUSE complex nucleic acid and protein bind on opposite sides of a two-domain structural unit (Fig. 4b) and that the two binding events are fully independent—e.g. the Kds measured in the bi-molecular complex (~10 μM for both interactions) do not vary in the presence of the third component. This indicates that, in the cell, the coupling between two key c-myc transcriptional regulators is mediated by a pure physical tethering and does not involve an allosteric effect (Cukier et al. 2010).
The study of multiple interactions by NMR. a Information on the interaction surfaces obtained on single domain constructs can be used to characterise protein-RNA interactions in larger constructs—schematic. b FIR RRM1-RRM2 (grey surface) binds the FBP protein and ssDNA molecule on two physically separated sites (purple and green, respectively), located on opposite sides of the molecule (PDB: 2KXF) (Cukier et al. 2010). c RDC-, PRE- and SAXS-derived information allows determining the orientation of different domains/molecules within large complexes
An important requirement for the analysis of multi-component protein-RNA complexes is the detection of non-overlapping NMR signal in complexes of high molecular weight. The use of line-narrowing experiments recorded at high or ultra-high fields combined with deuteration has allowed recording fingerprint spectra for protein-RNA complexes of >50 kDa (Garcia-Mayoral et al., unpublished, Oberstrass et al. 2005). New methodology where the use of RDC, PRE (and in some cases Pseudo Contact Shift) information is coupled to techniques that provide a molecular envelope (e.g. SAXS, SANS) have been successful in providing the structural description of multi-domain protein-RNA complexes of 30–40 kDa (Madl et al. 2010) (Fig. 4c). However, when coupled to segmental labelling techniques that reduce the complexity of the NMR spectra by selecting only a part of the molecule (Skrisovska et al. 2010), we expect these techniques to allow building structural models for much larger multi-component complexes. Interestingly, the 13C labelling of a protein’s methyl groups has been successful in the determination of the global fold of an 82-kDa protein (Tugarinov et al. 2005) as well as in answering specific questions in much larger (100–1,000 kDa) systems where previous structural information was available (Ruschak and Kay 2010). Such a labelling is likely to play a significant role in the probing of dynamics and interactions within high-molecular-weight protein-RNA complexes.
Above we describe some of the unique advantages of NMR in the study of transient protein-RNA complexes. We have focused on approaches of general use, although a number of more specific experiments can report on, for example, important protein side chain motions (Iwahara et al. 2007; Mulder et al. 2001). Further, ad hoc NMR experiments have allowed capturing scarcely populated protein conformers and transient protein-ligand complexes (Iwahara and Clore 2006; Tang et al. 2006). We can expect these experiments to be adapted to monitor protein-RNA interactions. Finally, in the next few years we can expect that continuous improvements in sensitivity and, importantly, the development of tools to streamline the preparation of segmentally labelled proteins will allow NMR to play a growing role in the molecular characterisation of mRNA metabolism.
References
Allain FH, Gubser CC, Howe PW, Nagai K, Neuhaus D, Varani G (1996) Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature 380:646–650
Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM (2007) A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins 69:449–465
Auweter SD, Oberstrass FC, Allain FH (2006) Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res 34:4943–4959
Bahadur RP, Zacharias M (2008) The interface of protein-protein complexes: analysis of contacts and prediction of interactions. Cell Mol Life Sci 65:1059–1072
Beuth B, Garcia-Mayoral MF, Taylor IA, Ramos A (2007) Scaffold-independent analysis of RNA-protein interactions: the Nova-1 KH3-RNA complex. J Am Chem Soc 129:10205–10210
Buckanovich RJ, Darnell RB (1997) The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol 17:3194–3201
Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG (1997) The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell 88:235–242
Clery A, Blatter M, Allain FH (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18:290–298
Conte MR, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J 19:3132–3141
Cukier CD, Hollingworth D, Martin SR, Kelly G, Diaz-Moreno I, Ramos A (2010) Molecular basis of FIR-mediated c-myc transcriptional control. Nat Struct Mol Biol 17:1058–1064
De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF (1998) Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388
Deka P, Rajan PK, Perez-Canadillas JM, Varani G (2005) Protein and RNA dynamics play key roles in determining the specific recognition of GU-rich polyadenylation regulatory elements by human Cstf-64 protein. J Mol Biol 347:719–733
Dominguez C, Schubert M, Duss O, Ravindranathan S, Allain FH (2011) Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 58:1–61
Doyle M, Jantsch MF (2003) New and old roles of the double-stranded RNA-binding domain. J Struct Biol 140:147–153
Ellis JJ, Broom M, Jones S (2007) Protein-RNA interactions: structural analysis and functional classes. Proteins 66:903–911
Garcia-Mayoral MF, Hollingworth D, Masino L, Diaz-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A (2007) The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure 15:485–498
Garcia-Mayoral MF, Diaz-Moreno I, Hollingworth D, Ramos A (2008) The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res 36:5290–5296
Hall TM (2005) Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15:367–373
Iwahara J, Clore GM (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440:1227–1230
Iwahara J, Jung YS, Clore GM (2007) Heteronuclear NMR spectroscopy for lysine NH(3) groups in proteins: unique effect of water exchange on (15)N transverse relaxation. J Am Chem Soc 129:2971–2980
Keene JD, Tenenbaum SA (2002) Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell 9:1161–1167
Kirkpatrick JP, Li P, Carlomagno T (2009) Probing mutation-induced structural perturbations by refinement against residual dipolar couplings: application to the U4 spliceosomal RNP complex. Chembiochem 10:1007–1014
Leulliot N, Quevillon-Cheruel S, Graille M, van Tilbeurgh H, Leeper TC, Godin KS, Edwards TE, Sigurdsson ST, Rozenkrants N, Nagel RJ, Ares M, Varani G (2004) A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J 23:2468–2477
Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456:464–469
Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294:1098–1102
Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, Smith CW (2010) Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17:1114–1123
Lunde BM, Moore C, Varani G (2007) RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8:479–490
Madl T, Gabel F, Sattler M (2010) NMR and small-angle scattering-based structural analysis of protein complexes in solution. J Struct Biol (In Press, Corrected Proof)
Maguire ML, Guler-Gane G, Nietlispach D, Raine AR, Zorn AM, Standart N, Broadhurst RW (2005) Solution structure and backbone dynamics of the KH-QUA2 region of the Xenopus STAR/GSG quaking protein. J Mol Biol 348:265–279
Mansfield KD, Keene JD (2009) The ribonome: a dominant force in co-ordinating gene expression. Biol Cell 101:169–181
Martin-Tumasz S, Reiter NJ, Brow DA, Butcher SE (2010) Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24. RNA 16:792–804
Mittermaier A, Varani L, Muhandiram DR, Kay LE, Varani G (1999) Changes in side-chain and backbone dynamics identify determinants of specificity in RNA recognition by human U1A protein. J Mol Biol 294:967–979
Mulder FA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE (2001) Measurement of slow (micros-ms) time scale dynamics in protein side chains by (15)N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J Am Chem Soc 123:967–975
Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH (2005) Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309:2054–2057
Pancevac C, Goldstone DC, Ramos A, Taylor IA (2010) Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3′-end processing factors. Nucleic Acids Res 38:3119–3132
Ramos A, Hollingworth D, Major SA, Adinolfi S, Kelly G, Muskett FW, Pastore A (2002) Role of dimerization in KH/RNA complexes: the example of Nova KH3. Biochemistry 41:4193–4201
Ravindranathan S, Oberstrass FC, Allain FH (2010) Increase in backbone mobility of the VTS1p-SAM domain on binding to SRE-RNA. J Mol Biol 396:732–746
Reid DC, Chang BL, Gunderson SI, Alpert L, Thompson WA, Fairbrother WG (2009) Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence. RNA 15:2385–2397
Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW (2006) A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat Struct Mol Biol 13:839–848
Ruschak AM, Kay LE (2010) Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR 46:75–87
Shajani Z, Drobny G, Varani G (2007) Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry 46:5875–5883
Skrisovska L, Schubert M, Allain FH (2010) Recent advances in segmental isotope labeling of proteins: NMR applications to large proteins and glycoproteins. J Biomol NMR 46:51–65
Tang C, Iwahara J, Clore GM (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444:383–386
Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE (2001) Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc 123:11341–11352
Treger M, Westhof E (2001) Statistical analysis of atomic contacts at RNA-protein interfaces. J Mol Recognit 14:199–214
Tugarinov V, Choy WY, Orekhov VY, Kay LE (2005) Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc Natl Acad Sci USA 102:622–627
Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science 302:1212–1215
Valverde R, Edwards L, Regan L (2008) Structure and function of KH domains. FEBS J 275:2712–2726
Varani L, Gunderson SI, Mattaj IW, Kay LE, Neuhaus D, Varani G (2000) The NMR structure of the 38 kDa U1A protein—PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol 7:329–335
Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH (2009) An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16:130–137
Acknowledgments
Open Access distribution of this review and the work of A. Ramos have been supported by the Medical Research Council Grant U117574558.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Transient interactions in biology.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Cukier, C.D., Ramos, A. Modular protein-RNA interactions regulating mRNA metabolism: a role for NMR. Eur Biophys J 40, 1317–1325 (2011). https://doi.org/10.1007/s00249-011-0698-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-011-0698-z