Abstract
Karst aquifers are a significant source of drinking water and highly vulnerable to pollution and microbial contamination. Microbiological regulations for the quality of drinking water mostly focus on bacterial levels and lack guidance concerning fungal contamination. Moreover, there is no standardised microbial analysis methodology for identifying fungi in water. Our main objective was to establish the most effective culture and identification methodology to examine yeast diversity in karst waters. We assessed the comparative efficacy of four culture media (CHROMagar Candida, dichloran glycerol 18% [DG18], dichloran rose Bengal chloramphenicol [DRBC], and SYMPHONY agar) for yeast isolation from karst water samples. Furthermore, we investigated the comprehensiveness of databases used in MALDI-TOF mass spectrometry (MALDI-TOF MS) for identifying environmental yeast species. In total, we analysed 162 water samples, allowing the identification of 2479 yeast isolates. We demonstrate that a combination of four culture media, each with distinct specifications, more efficiently covers a wide range of yeast species in karst water than a combination of only two or three. Supplementation of a MALDI-TOF MS database is also critical for analysing environmental microbial samples and improved the identification of yeast biodiversity. This study is an initial step towards standardising the analysis of fungal biodiversity in karst waters, enabling a better understanding of the significance of this environmental reservoir in relation to public health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Karst groundwater is a significant source of drinking water worldwide and is believed to serve approximately 25% of the world’s population [1]. Water of karst systems is particularly vulnerable to a wide range of chemical and biological pollutants, which have a direct impact on the quality of drinking water [2]. As a result, water from karst aquifers represents a potential risk of exposure to contaminants for the human population, and, therefore, drinking water regulations are essential. Currently, the microbiological analysis of drinking water focuses primarily on bacterial parameters that indicate faecal contamination. Drinking water safety and quality regulations are heavily based on the presence of Escherichia coli, enterococci, coliforms, and clostridia [3, 4]. The inclusion of fungi in current national or international drinking water regulations is practically non-existent, although their presence in water has been well established over the last 30 years [5, 6]. The Swedish National Food Administration is the only legislation that includes a parametric value for fungi in drinking water [7].
The literature on the biodiversity of fungi in karst waters is scarce compared to that on bacteria. Several recent studies have characterised the fungi found in drinking water, but most focused mainly on filamentous fungi, and little attention has been accorded to yeast in water [8,9,10]. These studies have used culture methods to fungi in water, using different isolation media with various incubation times and temperatures. The heterogeneity of isolation procedures has been highlighted previously [5, 10]. High nutrient media are often used, such as Sabouraud dextrose agar and malt extract agar, but are quickly covered by fast-growing moulds [8,9,10]. Alternatively, more selective media containing fungicides that inhibit overgrowth of these fast-growing moulds, such as dichloran-18% glycerol agar (DG18) or dichloran-Rose Bengal agar (DRBC) [6, 11], were used. The use of different media may result in selectivity for some fungal species and loss of others. Most of the studies used a single medium, sometimes two, and have mainly reported the presence of moulds in the water and, sporadically, of yeasts. [6, 11]. Thus, many aspects concerning the biodiversity of yeast in drinking water are still poorly understood. Currently, there are no international guidelines for the analysis of fungi in drinking water. The lack of a standardised protocol makes it difficult to compare studies and ensure reliable results due to significant differences in methodology [11]. To date, there is a lack of studies evaluating the ability of different culture media to characterise fungal biodiversity in water as effectively as possible, especially for yeasts.
There is an urgent need to define a standard protocol for the analysis of fungal biodiversity in water. The current study is part of the TRANSKARST project, a transdisciplinary study of karst waters in France that was launched in 2019. The primary endpoint of this study is to identify the best culture strategy for yeast in karst waters using a combination of four culture media. Second, we assessed the importance of complementing and combining existing MALDI-TOF MS databases to improve the identification of environmental yeasts. This has enabled us to identify a very high level of fungal biodiversity in these waters, which has not been previously reported.
Materials and Methods
Study Sites and Sample Collection
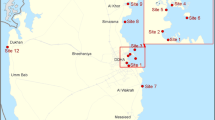
The study area is the recharge area of the Arcier spring (102 km2) located in the Jura Mountains (Eastern France). This spring (ARC; 47.2673538, 6.1209294) is the outlet of a complex karst system, extending mainly on the Saône plateau. Two main swallow holes allow surface water to enter the underground and contribute to groundwater circulation: the Creux-sous-Roche (CRE) and Nancray swallow holes (PER). Near the town of Saône (47.233333, 6.116667), the CRE drains the western part of the plateau, collecting water from (1) the Saône swamp (800 ha) and (2) the FSA stream via the PSC spring. The PER is located near the town of Nancray (47.2448, 6.1806) and drains the waters of the RUI spring, as well as the eastern part of the plateau. Mapping and schematic representation of the Arcier karst watershed are available elsewhere [12]. The Arcier spring is crucial in terms of water quality and quantity at the local scale because it is the main drinking water supply for the city of Besançon (118,000 inhabitants).
Water samples were collected in sterile Falcon flasks once or twice a month from June 2020 to November 2021 at each of the six selected sampling sites (ARC, CRE, FSA, PER, PSC, and RUI). Samples were transported at +4 °C and filtered on the day of collection. Due to logistical constraints, some sampling sessions could not be carried out. In seven sampling sessions, PSC was not sampled because of flooding.
Culture Media
Four different culture media were used during the study. Their choice was based on their capacity of isolation of yeast and/or limitation of filamentous overgrowth. DG18 and DRBC media (BIOKAR, Allonne, France) inhibit filamentous growth due to the presence of dichloran, as previously described [12]. CHROMagar Candida medium (Graso, Starogard Gdanski, Poland) facilitates the macromorphological differentiation of yeast colonies belonging to different species [13]. SYMPHONY medium (BIOKAR, Allonne, France) has been recently developed for the analysis of environmental yeasts and moulds and the analysis of membrane-filtered water. We evaluated the performance of each media alone or in combination of two or three, compared with the combination of four media. We arbitrarily considered that the combination of the four media allowed the development of all the yeasts.
Sample Filtration and Yeast Isolation
Yeast isolation procedure was based on the Swedish standard method for microfungal analysis in water [14]. Volumes of 100 mL of the water samples were filtered through 0.45 μm cellulose membranes (EZ-Fit™ filtration units, Millipore, Molsheim, France) by vacuum filtration. The membranes were then placed on 90 mm petri dishes with DG18, CHROMagar, DRBC, or SYMPHONY agar medium and incubated at 30 °C for 7 days. One sterile filtration unit was used per sampling location, and a different membrane was deposited on each agar plate. Yeast colonies were counted after 20 to 24 h (day 1) and 40 to 44 h (day 2) of incubation for the CHROMagar, DRBC, and SYMPHONY media and the plates then checked daily for new isolates until day 7 or complete overgrowth by moulds. For the DG18 media, the colony count was performed after incubation of 40 to 44 h (day 2) and 60 to 66 h (day 3), and the plates then checked for new isolates until day 7 or complete overgrowth by moulds. Macromorphological criteria allowed the differentiation of yeast colonies belonging to different species. Colony forming units (CFUs) were quantified and individual strains isolated and subcultured on fresh petri dishes with Sabouraud dextrose agar. These were incubated at 30 °C for 2 to 3 days, until identification. All yeast strains were stored in cryogenic vials at −80 °C.
MALDI-TOF Mass Spectrometry
Yeast identification was performed primarily by matrix-assisted laser desorption ionisation coupled to time-of-flight mass spectrometry (MALDI-TOF MS) using a Bruker microflex® LRF instrument (Bruker Daltonics, Bremen, Germany). Initially, each yeast colony was analysed using a rapid MALDI-TOF MS protocol with a rapid formic acid extraction performed on subcultures after 48 h of incubation. According to the protocol of the manufacturer, a thin layer of the colony was applied to a spot on the MALDI steel plate, followed by the addition of 1 μL formic acid (70%) and then 1 μL HCCA matrix, before starting the MALDI run. This short protocol was repeated if the previous one was unsuccessful. Subsequently, a long MALDI-TOF MS protocol with preliminary formic acid/acetonitrile protein extraction [15] was performed to analyse the samples that remained unidentified after the two short protocols, and repeated one time if necessary. Thus, two rapid protocols, followed by two long protocols, were performed before yeast isolates were considered as unidentified. Final spectral data were initially compared to the MSI-2 database (https://msi.happy-dev.fr/) and then to the Bruker Biotyper Compass reference library. According to the manufacturer’s instructions, species-level identification was considered if the Bruker library LogScore was ≥1,7 and/or if the MSI-2 database score was between 20 and 100. The strains were identified using rRNA single gene barcoding marker if there was a mismatch between the MSI-2 database and the Bruker library or if they failed to be identified by either MS database.
DNA Extraction
DNA was extracted from the yeast that remained unidentified or misidentified after the MALDI-TOF MS assays using the Chelex DNA extraction method [16]. A loopful of a yeast colony was suspended in 600 μL sterile water and 200 μL Chelex resin and incubated at 100 °C for 30 min. The yeast suspensions were then vortexed briefly and centrifuged at 14,000 rpm for 10 min. The supernatants were collected and diluted 1:10 in sterile water. A volume of 2 μL of DNA extract was used immediately for PCR. The remainder of the DNA extract was stored at −20 °C.
Amplification and Sequencing of rRNA Genes
DNA extracts were amplified by PCR in a 50-μL volume containing 2 μL 1:10 DNA extract, 5 μL each of 3 µmol/L primers, and 25 μL FastStart PCR Master Mix (Roche, Basel, Switzerland). The internal transcribed spacer (ITS) region of the ribosomal RNA (rRNA) gene was amplified using forward primer ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and reverse primer ITS4 (5′-GCATATCAATAAGCGGAGGA-3′). In the event of unsuccessful ITS amplification, the D1/D2 region of the 26S rRNA gene was amplified using universal primers, such as the NL1 forward primer (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 reverse primer (5′-GGTCCGTGTTTCAAGACGG-3′). PCR amplicons were separated by gel electrophoresis on 2% agarose gels in 1X Tris-acetate-EDTA (TAE), visualised using ethidium bromide under UV transillumination. Then, these were purified and diluted to obtain a DNA concentration between 20 and 40 μmol/L. DNA concentration was quantified on a NanoDrop instrument (Thermo Fisher Scientific, USA). PCR amplicons were sequenced using the reverse primer amongst ITS4 and NL4 at 10 µmol/L. Yeast identifications were obtained by first comparing the sequences using the Westerdijk Fungal Biodiversity Institute pairwise alignment (https://wi.knaw.nl/Pairwise_alignment) and then the GenBank Nucleotide BLAST database (https://blast.ncbi.nlm.nih.gov/Blast). According to the literature, the taxonomic threshold used for species level identification is 98.41% for the ITS and 99.51% for NL [17]. In the event that identification at the species level was unsuccessful, identification at the genus level was retained using a taxonomic threshold of 96.31% for the ITS and 97.11% for NL [17].
Unidentified Strains
“Lost strains” were strains that died before identification. “Unknown strains” were those that remained unidentified after both MALDI-TOF MS and PCR assays.
Supplementation of the MSI Database
The strains that were not identified by MALDI-TOF MS but successfully identified by PCR were thawed at room temperature and subcultured on both Sabouraud dextrose agar and CHROMagar. A long MALDI-TOF MS protocol, as previously described, was run on each strain from both CHROMagar and Sabouraud agar in quadruplicate. The spectral data obtained were assigned to the appropriate species and then submitted to the MSI-2 database curator for inclusion.
Filamentous Growth Quantification
Filamentous colonies were quantified twice daily (respectively after 24, 42, 48, 66, 72, 90, 96, 120, 144 et 168 hours of incubation) during the seven-day incubation period. Each primary agar plate was assigned a percentage cellulose membrane invasion value relative to a clean membrane surface.
Statistics
Ordinary one-way analysis of variance (ANOVA) tests followed by Tukey’s multiple comparison test were used to compare the performance of the media combinations. The ⍺ value was set to 0.05. Statistical analysis was performed, and the figure generated using GraphPad Prism version 9.5.0 (GraphPad Software, Boston, USA).
Results
In total, 162 water samples were collected during the study, representing 29 sampling sessions. All the seasons were covered to obtain the best possible representation of biodiversity, which is potentially influenced by physical and chemical factors, depending on the climatic condition. A total of 2479 yeast isolates were analysed during the study, corresponding to 1485 identified strains, with an average of 9.2 strains identified per sample.
Biodiversity
From a pool of 2479 yeast isolates analysed during the study, 2389 isolates (96.4%), representing 1485 strains, were identified. Among them, 2339 isolates, representing 1449 strains, were identified at the species level, and belonged to 142 different species. The remaining 50 isolates, representing 36 strains, were identified at the genus level and belonged to 10 different genera. There was a large disparity in the number of strains of each species. Indeed, three species (Pichia fermentans, Metschnikowia pulcherrima, Hanseniaspora uvarum) accounted for 25% of all strains identified and 30 for almost 75% of all isolated strains (Table 1). A list with all the species identified during the study is given in supplementary material (Table S1).
Ninety isolates (3.6%) remained unidentified at the end of the study. The lost strains (n=65) were mostly unidentifiable due to early overgrowth by filamentous fungi. The unknown strains (n=25), corresponding to strains that could not be identified using molecular barcoding, appeared to be evenly distributed among the four media (range from four to eight). These 90 isolates represented an undetermined number of species and were not considered in the evaluation of the performance of the media.
Performance of the Media
Evaluation of the media’s performance was based on the 152 species identified by the combination of the four media. An average of 90/152 species (58.9%) were found using only one culture medium for yeast identification. The DRBC and SYMPHONY media were the most effective, with 97 (63.8%) species potentially isolated, whereas the DG medium was the least effective, with 73 (48%) species isolated (Fig. 1). The average number of species using two media was 118/152 (78.1%), ranging from 107 to 126 (70.4–82.9%). Using a combination of three media was more effective, with an average of 138/152 (90.6%) species isolated, ranging from 133 to 144 (87.5–94.7%) (Table 2).
Using a four-media combination was significantly more efficient for species identification than using a single media (p<0.0001) or a combination of two media (p=0.0009). The four-media combination was more efficient than the three-media combinations, but the difference was not statistically significant (p=0.1955). The four-media combination also allowed a significantly higher number of isolates to be identified than either three-media (p=0.0002) or two-media (p<0.0001) combinations.
Invasion of the Culture Media
On average, 35% of the surface of the CHROMagar and SYMPHONY media was overgrown with filamentous fungi after 40 h of incubation versus 13% of the DRBC surface and only 3% of that of DG18. After 66 h of incubation, nearly 90% of the surface of the CHROMagar and SYMPHONY media was overgrown versus 65% of the DRBC surface and less than 50% of that of DG18. After 96 h of incubation, all media were considered to be 100% overgrown (Fig. 2).
MALDI-TOF MS vs PCR
MALDI-TOF MS allowed the identification of 2119 isolates (88.7%), representing 113 of 152 species (74.3%). The rest of the isolates (n=270; 11.3%) were additionally identified by sequencing. They belonged to 88 different species, of which 39, representing 77 isolates, were identified only by sequencing analysis.
During the study, we added spectral data for 21 species to the MSI-2 database. Among them, 15 were new species that were missing from the original database. The other six were complementary spectra added to species already existing in the database. Six of these 21 species were among the 30 most common species that we identified (Table 1). At the end of our study, we compared the initial databases (June 2020) to those that had been supplemented (November 2021). In June 2020, 65 species of the 152 we identified during the study were identifiable with the initial MSI-2 database, corresponding to 62.7% of our isolates. After the addition of new species, the supplemented MSI-2 database (November 2021) could identify 95 species, corresponding to 88% of our isolates. The use of supplemented MSI-2 database combined with the Bruker library allowed the identification of 2283 isolates (92%) from 111 species (73%).
Discussion
Combining Culture Media Enhances the Biodiversity of Yeast Identified in Water
Culture-based methods are low-cost, practical in routine practise, and do not require a high level of expertise [5]. The use of multiple and diverse combinations of media has already been shown to increase the biodiversity of fungi found in water [10]. However, yeasts have rarely been studied specifically. This led us to investigate a strategy combining a medium specifically developed for yeasts (CHROMagar Candida), another nutrient-rich medium (Symphony), and two media containing dichloran, known to inhibit the growth of fast-growing moulds. We found the identification of yeast biodiversity in karst water to be significantly enhanced by using a combination of complementary media. While a single culture medium only detects 59% of biodiversity, combinations of media improve performance, detecting 78% and 91% of biodiversity, respectively with two or three media. The four-media combination was chosen as the standard with an arbitrary optimum performance of 100%. However, this does not reflect reality. While the four-environment strategy provides optimal biodiversity, we cannot be certain that all cultivable species have been isolated using this combination. The use of other additional media could have further increased biodiversity, reducing the relative performance of the four-media combination.
In this study, the combination of media enabled the coverage of a wide range of species due to their specific properties. Due to its chromogenic detection, CHROMagar Candida medium enhances the detection of yeast with specific enzymatic activity [18]. Hence, it facilitates the differentiation and isolation of yeast colonies on primary culture plates based on macromorphological criteria. CHROMagar medium is particularly efficient for the detection of Candida species, Pichia species, and Rhodotorula species [18, 19]. Indeed, we observed these species to be highly present in our karst water samples using this medium (Table 1). Thus, CHROMagar with DRBC was the best two-media combination for yeast recovery in our study. At least, using a chromogenic medium such as CHROMagar for the analysis of karst water avoids duplicate isolation of the same strains and is cost-effective while still allowing the detection of high yeast biodiversity.
The DG18 and DRBC media are selective for the identification of yeast by inhibiting filamentous fungi overgrowth due to the presence of dichloran. DRBC medium has been widely used for yeast identification in water samples [14, 20,21,22,23] since it was reported in the Swedish standard method for the analysis of micro-fungi in water [14]. According to the literature and our results, DRBC medium is the most efficient for the identification of high yeast biodiversity [21]. However, we found DG18 medium to be more efficient in inhibiting filamentous growth and its capacity to isolate xerophilic species and black yeast-like species of interest [24, 25]. The DG18 medium allowed us to isolate five of six strains of Aureobasidium pullulans group. Of note, MALDI-TOF MS database did not allow differential diagnosis between A. pullulans and its sister species A. melanogenum, previously described with high prevalence in Slovenian water [26]. Moreover, DG18 medium promoted growth of 32 out of 52 strains of the xerophilic species Debaryomyces hansenii. Although the DG18 medium showed the lowest identification rate in our study, it was still essential for the identification of certain species and enlarging the yeast biodiversity identified.
SYMPHONY medium has only been recently developed for food analysis. There are currently no studies in the literature on this medium. However, it appears to be promising for the analysis of yeast in water, as we found it to be as efficient as DRBC medium for yeast diversity identification (Fig. 1). It also allowed the identification of a strain belonging to the black-yeast Aureobasidium pullulans group. Further studies are needed to evaluate this medium in environmental studies.
Supplementing and Combining the MALDI-TOF MS Databases Is Essential
MALDI-TOF MS has recently emerged as an alternative to conventional historical biochemical tests for the routine clinical identification of yeast. This transition has been attributed to its higher efficiency, rapid processing, and cost-effectiveness [27, 28]. Although MALDI-TOF MS is widely used in mycology laboratories for the identification of pathogenic species, its applicability to environmental species has been hampered by substantial gaps in existing databases. As a result, MALDI-TOF MS is largely underrepresented in studies of environmental yeast biodiversity. In the literature, the identification of environmental yeast species is mainly performed by molecular approaches based on PCR and sequencing targeting the ITS region [6, 29,30,31,32]. However, the use of MALDI-TOF MS allows significant savings in terms of both time and cost [33,34,35]. Consequently, adopting MALDI-TOF MS over PCR and sequencing methods offers the opportunity to analyse a substantially larger number of isolates.
Supplementing the databases with the spectra of missing species is essential to increasing the potential environmental applications of MALDI-TOF MS [36]. Our strategy consisted of supplementing the MSI-2 database because it has the advantage of being a collaborative and open access database. At the end of our study, the supplemented MSI-2 database was able to identify 88% of our isolates versus 63% at the beginning, and the number of identifiable species increased from 65 to 95 out of the 152 submitted. This is a significant improvement in the identification rate of environmental yeast biodiversity in water and demonstrates the importance of supplementing the databases. Furthermore, the addition of spectra from 21 species to the MSI-2 database allowed us to include 315 of our isolates, 123 belonging to the five species that were among the 30 most frequently observed. On the contrary, the updated Bruker library contained only three new species that we identified, representing 29 isolates. This demonstrates the effectiveness of supplementing a freely accessible database, such as MSI-2. However, the best way to improve the identification of environmental yeasts remains the combination of databases. Indeed, combining the MSI-2 database with the Bruker library allowed us to identify 111 species, representing 92% of our isolates. The databases are complementary: combining them improves the ability to identify more yeast species as well as the confidence level of identification. At last, adding new spectra from different strains of the same species improves the identification rate and provides better taxonomic identification scores [36].
A limitation of our study was that we compared the databases retrospectively. During the study, we were unable to supplement the MSI-2 database in real time. A few weeks elapsed between the molecular identification of a newly identified species and the acquisition of its spectra in the database. Therefore, we could only retrospectively estimate the performance of the database at the beginning of our study and compare it to the supplemented one at the end of our study.
Importance of the Microbial Analysis of Drinking Water
Human-made environments, such as households and hospitals, are known to be major habitats for opportunistic fungi [37]. Groundwater from karst aquifers favours the transmission of fungal contaminants to drinking water, and numerous studies have identified pathogenic fungi in household water systems [5, 6, 31, 38, 39]. In our study, several opportunistic pathogenic yeast species were identified, such as Candida parapsilosis, Rhodotorula mucilaginosa, and Exophiala dermatitidis [40, 41]. These species have been reported to be emerging pathogens and have already been identified in groundwater and households, representing a source of human exposure. In addition, we found only several strains of the human pathogen Candida albicans, which may be an indicator of human contamination of drinking water sources. We also reported the high presence of Nakaseomyces glabratus (formerly Candida glabrata), the second most common human pathogen isolated from yeast fungemia [42]. Furthermore, we observed the high presence of Pichia kudriavzevii (formerly Candida krusei), which is intrinsically fluconazole resistant and also frequently associated with human fungemia. With an increasing immunosuppressed population, it is crucial to analyse yeast biodiversity in environmental sources of drinking water, such as karst waters. Our current knowledge of the biodiversity of environmental yeast species and their transmission by drinking water reveals the need to regulate their presence in drinking water. Thus, the development of a standard analytical protocol is crucial.
Conclusion
In this study, our strategy was to improve the identification of yeast biodiversity in karst waters by developing an optimal culture-based method. The number of strains identified in our study was much higher than in any other study in the literature, which is an encouraging and promising result. Moreover, our work clearly demonstrates the importance of using complementary media to study the diversity of yeasts in water and justifies at least a combination of a nutrient-rich medium and a selective medium containing dichloran. The use of MALDI-TOF MS, combined with improved databases enriched with environmental strains, enables further larger-scale studies to be carried out at a lower cost. However, it only allows the species present in the base to be identified. The drawback of the MS databases justifies the complementary use of DNA barcoding to identify species that cannot be identified or distinguished using MALDI-TOF MS. As drinking water is an important source of human exposure to fungal contaminants, its analysis needs to continue. The analysis of yeast biodiversity in karst waters is a first step towards assessing the biosafety level of each fungal contaminant and establishing drinking water regulations. The next step will be to analyse many drinking water networks, identify their yeast biodiversity, and investigate the parameters that influence it.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. Supplementary material associated with this article can be found in the online version.
References
Ford D, Williams P (2007) Karst hydrogeology and geomorphology: Ford/Karst Hydrogeology and Geomorphology. John Wiley & Sons Ltd, West Sussex, England
White WB (2018) Contaminant transport in karst aquifers: systematics and mechanisms. In: White WB, Herman JS, Herman EK, Rutigliano M (eds) Karst groundwater contamination and public health. Springer International Publishing, Cham, pp 55–81
World Health Organization (2017) Guidelines for drinking-water quality: fourth edition incorporating first addendum, 4th ed + 1st add. World Health Organization, Geneva
Union E (2020) Directive (EU) 2020/2184 of the European Parliament and of the Council of 16 December 2020 on the quality of water intended for human consumption. Off J Eur Union 435:1–62
DEFRA (Department for Environment, Food & Rural Affairs) (2011) A review of fungi in drinking water and the implications for human health1st edn. BIO Intelligence Service https://www.dwi.gov.uk/research/completed-research/risk-assessment-microbiological/a-review-of-fungi-in-drinking-water-and-the-implications-for-human-health/. Accessed 22 Aug 2023
Babič M, Gunde-Cimerman N, Vargha M et al (2017) Fungal contaminants in drinking water regulation? A tale of ecology, Exposure, purification and clinical relevance. Int J Environ Res Public Health 14:636. https://doi.org/10.3390/ijerph14060636
Administration SNF (2022) “Livsmedelsverkets föreskrifter om dricksvatten” (in Swedish). LIVSFS 2022:12 https://www.livsmedelsverket.se/foretagande-regler-kontroll/dricksvattenproduktion/regler-om-dricksvatten. Accessed 11 Aug 2023
Kanzler D, Buzina W, Paulitsch A et al (2008) Occurrence and hygienic relevance of fungi in drinking water. Mycoses 51:165–169. https://doi.org/10.1111/j.1439-0507.2007.01454.x
Sammon NB, Harrower KM, Fabbro LD, Reed RH (2010) Incidence and distribution of microfungi in a treated municipal water supply system in Sub-Tropical Australia. Int J Environ Res Public Health 7:1597–1611. https://doi.org/10.3390/ijerph7041597
Kinsey GC, Paterson RR, Kelley J (1998) Methods for the determination of filamentous fungi in treated and untreated waters. J Appl Microbiol 85:214S–224S. https://doi.org/10.1111/j.1365-2672.1998.tb05301.x
Hageskal G, Lima N, Skaar I (2009) The study of fungi in drinking water. Mycol Res 113:165–172. https://doi.org/10.1016/j.mycres.2008.10.002
Reboux G, Roussel S, Grenouillet F (2006) Moisissures de l’environnement agricole. J Mycol Med 16:248–262. https://doi.org/10.1016/j.mycmed.2006.09.003
Delhaes L, Touati K, Faure-Cognet O et al (2019) Prevalence, geographic risk factor, and development of a standardized protocol for fungal isolation in cystic fibrosis: results from the international prospective study “MFIP”. J Cyst Fibros 18:212–220. https://doi.org/10.1016/j.jcf.2018.10.001
Standard - Vattenundersökningar - Mikrosvampar i vatten - Kvantitativ bestämning med membranfiltermetod SS 28192. In: Svenska institutet för standarder, SIS. https://www.sis.se/produkter/naturvetenskap-och-tillampad-vetenskap/mikrobiologi/vattnets-mikrobiologi/ss28192/. Accessed 22 Aug 2023
Cassagne C, Cella A-L, Suchon P et al (2013) Evaluation of four pretreatment procedures for MALDI-TOF MS yeast identification in the routine clinical laboratory. Med Mycol 51:371–377. https://doi.org/10.3109/13693786.2012.720720
Provost F, Laurent F, Uzcategui LR, Boiron P (1997) Molecular study of persistence of Nocardia asteroides and Nocardia otitidiscaviarum strains in patients with long-term nocardiosis. J Clin Microbiol 35:1157–1160. https://doi.org/10.1128/jcm.35.5.1157-1160.1997
Vu D, Groenewald M, Szöke S et al (2016) DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol 85:91–105. https://doi.org/10.1016/j.simyco.2016.11.007
San-Millán R, Ribacoba L, Pontón J, Quindós G (1996) Evaluation of a commercial medium for identification of Candida species. Eur J Clin Microbiol Infect Dis 15:153–158. https://doi.org/10.1007/BF01591489
Pincus DH, Orenga S, Chatellier S (2007) Yeast identification—past, present, and future methods. Med Mycol 45:97–121. https://doi.org/10.1080/13693780601059936
Pereira VJ, Basílio MC, Fernandes D et al (2009) Occurrence of filamentous fungi and yeasts in three different drinking water sources. Water Res 43:3813–3819. https://doi.org/10.1016/j.watres.2009.05.024
Pereira VJ, Fernandes D, Carvalho G et al (2010) Assessment of the presence and dynamics of fungi in drinking water sources using cultural and molecular methods. Water Res 44:4850–4859. https://doi.org/10.1016/j.watres.2010.07.018
Turchetti B, Goretti M, Branda E et al (2013) Influence of abiotic variables on culturable yeast diversity in two distinct Alpine glaciers. FEMS Microbiol Ecol 86:327–340. https://doi.org/10.1111/1574-6941.12164
Gunde-Cimerman N, Zalar P, Hoog S et al (2000) Hypersaline waters in salterns—natural ecological niches for halophilic black yeasts. FEMS Microbiol Ecol 32:235–240. https://doi.org/10.1111/j.1574-6941.2000.tb00716.x
Hocking AD, Pitt JI (1980) Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl Environ Microbiol 39:488–492. https://doi.org/10.1128/aem.39.3.488-492.1980
Zalar P, Gostinčar C, De Hoog GS et al (2008) Redefinition of Aureobasidium pullulans and its varieties. Stud Mycol 61:21–38. https://doi.org/10.3114/sim.2008.61.02
Gostinčar C, Ohm RA, Kogej T et al (2014) Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics 15:549. https://doi.org/10.1186/1471-2164-15-549
Marklein G, Josten M, Klanke U et al (2009) Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol 47:2912–2917. https://doi.org/10.1128/JCM.00389-09
Sow D, Fall B, Ndiaye M et al (2015) Usefulness of MALDI-TOF mass spectrometry for routine identification of Candida species in a resource-poor setting. Mycopathologia 180:173–179. https://doi.org/10.1007/s11046-015-9905-2
Almeida JMGCF (2005) Yeast community survey in the Tagus estuary. FEMS Microbiol Ecol 53:295–303. https://doi.org/10.1016/j.femsec.2005.01.006
Gadanho M, Sampaio JP (2005) Occurrence and diversity of yeasts in the Mid-Atlantic ridge hydrothermal fields near the Azores archipelago. Microb Ecol 50:408–417. https://doi.org/10.1007/s00248-005-0195-y
Novak Babič M, Zalar P, Ženko B et al (2016) Yeasts and yeast-like fungi in tap water and groundwater, and their transmission to household appliances. Fungal Ecol 20:30–39. https://doi.org/10.1016/j.funeco.2015.10.001
Milanezi ACM, Witusk JPD, Van Der Sand ST (2019) Antifungal susceptibility of yeasts isolated from anthropogenic watershed. An Acad Bras Cienc 91:e20170369. https://doi.org/10.1590/0001-3765201820170369
Pliakos EE, Andreatos N, Shehadeh F et al (2018) The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev 31:e00095–e00017. https://doi.org/10.1128/CMR.00095-17
Zhao Y, Tsang C-C, Xiao M et al (2018) Yeast identification by sequencing, biochemical kits, MALDI–TOF MS and rep-PCR DNA fingerprinting. Med Mycol 56:816–827. https://doi.org/10.1093/mmy/myx118
Tran A, Alby K, Kerr A et al (2015) Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:2473–2479. https://doi.org/10.1128/JCM.00833-15
Agustini BC, Silva LP, Bloch C et al (2014) Evaluation of MALDI-TOF mass spectrometry for identification of environmental yeasts and development of supplementary database. Appl Microbiol Biotechnol 98:5645–5654. https://doi.org/10.1007/s00253-014-5686-7
Méheust D, Le Cann P, Reboux G et al (2014) Indoor fungal contamination: health risks and measurement methods in hospitals, homes and workplaces. Crit Rev Microbiol 40:248–260. https://doi.org/10.3109/1040841X.2013.777687
Döğen A, Sav H, Gonca S et al (2017) Candida parapsilosis in domestic laundry machines. Med Mycol 55:813–819. https://doi.org/10.1093/mmy/myx008
Döğen A, Kaplan E, Öksüz Z et al (2013) Dishwashers are a major source of human opportunistic yeast-like fungi in indoor environments in Mersin, Turkey. Med Mycol 51:493–498. https://doi.org/10.3109/13693786.2012.738313
Miceli MH, Díaz JA, Lee SA (2011) Emerging opportunistic yeast infections. Lancet Infect Dis 11:142–151. https://doi.org/10.1016/S1473-3099(10)70218-8
Kirchhoff L, Olsowski M, Rath P-M, Steinmann J (2019) Exophiala dermatitidis : key issues of an opportunistic fungal pathogen. Virulence 10:984–998. https://doi.org/10.1080/21505594.2019.1596504
Bretagne S, Sitbon K, Desnos-Ollivier M et al (2022) Active surveillance program to increase awareness on invasive fungal diseases: the French RESSIF network (2012 to 2018). mBio 13:e00920-22. https://doi.org/10.1128/mbio.00920-22
Acknowledgements
The authors are grateful to Elsa Lisciotto, Elodie Boucard, Céline Schmittlin, Margaux Hueber, Eva Ganard, and Brice Moulari for their valuable help in sample analysis.
Funding
This work was supported by the ISITE-BFC (Initiatives Science Innovation Territoire Economie en Bourgogne-Franche-Comté) through the SENSAAS (SENSors and Analyses for AquiferS) ISITE-BFC project (grant number: FC21010.CHR.IS—SENSAAS), by the Burgundy Franche-Comté region and the Agence de l’Eau Rhône-Méditerranée Corse, through the TRANSKARST (TRANSdisciplinary research on KARSTic waters) project (grant number: CRBFC: 2019-Y-09074), and the Zone Atelier de l’Arc Jurassien (IR RZA) and by Clinical Research and Innovation Division (DRCI) of Besançon University Hospital (CELIA grant).
Author information
Authors and Affiliations
Contributions
HC, XB, and FG contributed to the study conception and design. The sample collection was performed by VK, HC, and XB. The identification of the isolates and data collection were performed by FG, EP, and CG. ACN performed the addition of new spectra to the MSI database. The first draft of the manuscript was written by EP and CG. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Supplementary Information
ESM 1
(PDF 557 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grandhay, C., Prétot, E., Klaba, V. et al. Yeast Biodiversity of Karst Waters: Interest of Four Culture Media and an Improved MALDI-TOF MS Database. Microb Ecol 87, 26 (2024). https://doi.org/10.1007/s00248-023-02336-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00248-023-02336-1