Abstract
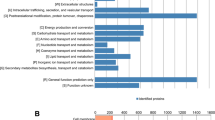
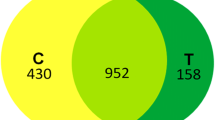
Trebouxia sp. (TR9) and Coccomyxa simplex (Csol) are desiccation-tolerant lichen microalgae with different adaptive strategies in accordance with the prevailing conditions of their habitats. The remodelling of cell wall and extracellular polysaccharides depending on water availability are key elements in the tolerance to desiccation of both microalgae. Currently, there is no information about the extracellular proteins of these algae and other aero-terrestrial microalgae in response to limited water availability. To our knowledge, this is the first report on the proteins associated with the extracellular polymeric substances (EPS) of aero-terrestrial microalgae subjected to cyclic desiccation/rehydration. LC-MS/MS and bioinformatic analyses of the EPS-associated proteins in the two lichen microalgae submitted to four desiccation/rehydration cycles allowed the compilation of 111 and 121 identified proteins for TR9 and Csol, respectively. Both sets of EPS-associated proteins shared a variety of predicted biological functions but showed a constitutive expression in Csol and partially inducible in TR9. In both algae, the EPS-associated proteins included a number of proteins of unknown functions, some of which could be considered as small intrinsically disordered proteins related with desiccation-tolerant organisms. Differences in the composition and the expression pattern between the studied EPS-associated proteins would be oriented to preserve the biochemical and biophysical properties of the extracellular structures under the different conditions of water availability in which each alga thrives.





Similar content being viewed by others
References
Costa OYA, Raaijmakers JM, Kuramae EE (2018) Microbial extracellular polymeric substances: ecological function and impact on soil aggregation. Front Microbiol 9:1636. https://doi.org/10.3389/fmicb.2018.01636
Vincent D, Rafiqi M, Job D (2020) The multiple facets of plant-fungal interactions revealed through plant and fungal secretomics. Front Plant Sci 10:1626. https://doi.org/10.3389/fpls.2019.01626
Krause C, Richter S, Knoll C, Jurgens G (2013) Plant secretome - from cellular process to biological activity. Biochim Biophys Acta 1834(11):2429–2441. https://doi.org/10.1016/j.bbapap.2013.03.024
Delaunois B, Jeandet P, Clément C, Baillieul F, Dorey S, Cordelier S (2014) Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front Plant Sci 5:249. https://doi.org/10.3389/fpls.2014.00249
Tanveer T, Shaheen K, Parveen S, Kazi AG, Ahmad P (2014) Plant secretomics: identification, isolation, and biological significance under environmental stress. Plant Signal Behav 9(8):e29426. https://doi.org/10.4161/psb.29426
Chen P, Jung NU, Giarola V, Bartels D (2020) The dynamic responses of cell walls in resurrection plants during dehydration and rehydration. Front Plant Sci 10:1698. https://doi.org/10.3389/fpls.2019.01698
Pierre G, Delattre C, Dubessay P, Jubeau S, Vialleix C, Cadoret JP, Probert I, Michaud P (2019) What is in store for EPS microalgae in the next decade? Molecules 24(23):4296. https://doi.org/10.3390/molecules24234296
Xiao R, Zheng Y (2016) Overview of microalgal extracellular polymeric substances (EPS) and their applications. Biotechnol Adv 34(7):1225–1244. https://doi.org/10.1016/j.biotechadv.2016.08.004
Chiou Y, Hsieh M, Yeh H (2010) Effect of algal extracellular polymer substances on UF membrane fouling. Desalination 250:648–652. https://doi.org/10.1016/j.desal.2008.02.043
Wang XQ, Yang PF, Liu Z, Liu WZ, Hu Y, Chen H, Kuang TY, Pei ZM, Shen SH, He YK (2009) Exploring the mechanism of Physcomitrella patens desiccation tolerance through a proteomic strategy. Plant Physiol 149(4):1739–1750. https://doi.org/10.1104/pp.108.131714
Geun Goo B, Baek G, Jin Choi D, Il Park Y, Synytsya A, Bleha R, Ho Seong D, Lee C, Kweon Park J (2013) Characterization of a renewable extracellular polysaccharide from defatted microalgae Dunaliella tertiolecta. Bioresour Technol 129:343–350. https://doi.org/10.1016/j.biortech.2012.11.077
Baba M, Suzuki I, Shiraiwa Y (2011) Proteomic analysis of high-CO(2)-inducible extracellular proteins in the unicellular green alga, Chlamydomonas reinhardtii. Plant Cell Physiol 52(8):1302–1314. https://doi.org/10.1093/pcp/pcr078
Terauchi M, Yamagishi T, Hanyuda T, Kawai H (2017) Genome-wide computational analysis of the secretome of brown algae (Phaeophyceae). Mar Genomics 32:49–59. https://doi.org/10.1016/j.margen.2016.12.002
Ruíz-May E, Sorensen I, Fei Z, Zhang S, Domozych DS, Rose JKC (2018) The secretome and N-glycosylation profiles of the Charophycean green alga, Penium margaritaceum, resemble those of Embryophytes. Proteomes 6(2). https://doi.org/10.3390/proteomes6020014
Kranner I, Beckett R, Hochman A, Nash TH (2008) Desiccation-tolerance in lichens: a review. Bryologist 111:576–594. https://doi.org/10.1639/0007-2745-111.4.576
Hell AF, Gasulla F, Gonzáez-Hourcade MA, del Campo EM, Centeno DC, Casano LM (2019) Tolerance to cyclic desiccation in lichen microalgae is related to habitat preference and involves specific priming of the antioxidant system. Plant Cell Physiol 60(8):1880–1891. https://doi.org/10.1093/pcp/pcz103
González-Hourcade M, Braga MR, del Campo EM, Ascaso C, Patino C, Casano LM (2020a) Ultrastructural and biochemical analyses reveal cell wall remodelling in lichen-forming microalgae submitted to cyclic desiccation-rehydration. Ann Bot 125(3):459–469. https://doi.org/10.1093/aob/mcz181
Casano LM, Braga MR, Álvarez R, del Campo EM, Barreno E (2015) Differences in the cell walls and extracellular polymers of the two Trebouxia microalgae coexisting in the lichen Ramalina farinacea are consistent with their distinct capacity to immobilize extracellular Pb. Plant Sci 236:195–204. https://doi.org/10.1016/j.plantsci.2015.04.003
González-Hourcade M, del Campo EM, Braga MR, Salgado A, Casano LM (2020b) Disentangling the role of extracellular polysaccharides in DT in lichen-forming microalgae. First evidence of sulfated polysaccharides and ancient sulfotransferase genes. Environ Microbiol (In press) 22:3096–3111. https://doi.org/10.1111/1462-2920.15043
Centeno DC, Hell AF, Braga MR, del Campo EM, Casano LM (2016) Contrasting strategies used by lichen microalgae to cope with desiccation-rehydration stress revealed by metabolite profiling and cell wall analysis. Environ Microbiol 18(5):1546–1560. https://doi.org/10.1111/1462-2920.13249
Oliveira P, Martins NM, Santos M, Couto NA, Wright PC, Tamagnini P (2015) The Anabaena sp. PCC 7120 exoproteome: taking a peek outside the box. Life (Basel) 5(1):130–163. https://doi.org/10.3390/life5010130
Casano LM, del Campo EM, García-Breijo FJ, Reig-Arminana J, Gasulla F, Del Hoyo A, Guéra A, Barreno E (2011) Two Trebouxia algae with different physiological performances are ever-present in lichen thalli of Ramalina farinacea. Coexistence versus competition? Environ Microbiol 13(3):806–818. https://doi.org/10.1111/j.1462-2920.2010.02386.x
Malavasi V, Skaloud P, Rindi F, Tempesta S, Paoletti M, Pasqualetti M (2016) DNA-based taxonomy in ecologically versatile microalgae: a re-evaluation of the species concept within the coccoid green algal genus Coccomyxa (Trebouxiophyceae, Chlorophyta). PLoS One 11(3):e0151137. https://doi.org/10.1371/journal.pone.0151137
Bold HC, Parker BC (1962) Some supplementary attributes in the classification of Chlorococcum species. Arch Mikrobiol 42:267–288. https://doi.org/10.1007/bf00422045
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57(1):289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Nielsen H, Tsirigos KD, Brunak S, von Heijne G (2019) A brief history of protein sorting prediction. Protein J 38(3):200–216. https://doi.org/10.1007/s10930-019-09838-3
Almagro-Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37(4):420–423. https://doi.org/10.1038/s41587-019-0036-z
Hiller K, Grote A, Scheer M, Munch R, Jahn D (2004) PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res 32(Web Server issue):W375–W379. https://doi.org/10.1093/nar/gkh378
Kall L, Krogh A, Sonnhammer EL (2007) Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res 35(Web Server issue):W429–W432. https://doi.org/10.1093/nar/gkm256
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64(3):643–651. https://doi.org/10.1002/prot.21018
Lum G, Meinken J, Orr J, Frazier S, Min XJ (2014) PlantSecKB: the plant secretome and subcellular proteome knowledgebase. Comput Mol Biol 4(1):1–17. https://doi.org/10.5376/cmb.2014.04.0001
Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, López R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Stothard P (2000) The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28(6):1102–1104. https://doi.org/10.2144/00286ir01
Herburger K, Holzinger A (2015) Localization and quantification of callose in the streptophyte green algae Zygnema and Klebsormidium: correlation with desiccation tolerance. Plant Cell Physiol 56:2259–2270. https://doi.org/10.1093/pcp/pcv139
Herburger K, Ryan LM, Popper ZA, Holzinger A (2018) Localisation and substrate specificities of transglycanases in charophyte algae relate to development and morphology. J Cell Sci 131:jcs203208. https://doi.org/10.1242/jcs.203208
Yuan JS, Yang X, Lai J, Lin H, Cheng ZM, Nonogaki H, Chen F (2007) The endo-beta-mannanase gene families in Arabidopsis, rice, and poplar. Funct Integr Genomics 7(1):1–16. https://doi.org/10.1007/s10142-006-0034-3
Rosseto FR, Manzine LR, de Oliveira NM, Polikarpov I (2016) Biophysical and biochemical studies of a major endoglucanase secreted by Xanthomonas campestris pv. campestris. Enzym Microb Technol 91:1–7. https://doi.org/10.1016/j.enzmictec.2016.05.007
Stone BA (2009) Chemistry of β-Glucans. In: Bacic A, Fincher GB, Stone BA (eds) Chemistry, biochemistry, and biology of 1-3 beta glucans and related polysaccharides. Academic Press, pp 5–46
Zhu J, Álvarez S, Marsh EL, Lenoble ME, Cho IJ, Sivaguru M, Chen S, Nguyen HT, Wu Y, Schachtman DP, Sharp RE (2007) Cell wall proteome in the maize primary root elongation zone. II Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol 145(4):1533–1548. https://doi.org/10.1104/pp.107.107250
Ye T, Shi H, Wang Y, Chan Z (2015) Contrasting changes caused by drought and submergence stresses in bermudagrass (Cynodon dactylon). Front Plant Sci 6:951. https://doi.org/10.3389/fpls.2015.00951
Li P, Zhang Y, Wu X, Liu Y (2018) Drought stress impact on leaf proteome variations of faba bean (Vicia faba L.) in the Qinghai-Tibet Plateau of China. 3 Biotech 8(2):110-018-1088-3. https://doi.org/10.1007/s13205-018-1088-3
Carmo LST, Martins ACQ, Martins CCC, Passos MAS, Silva LP, Araujo ACG, Brasileiro ACM, Miller RNG, Guimaraes PM, Mehta A (2019) Comparative proteomics and gene expression analysis in Arachis duranensis reveal stress response proteins associated to drought tolerance. J Proteome 192:299–310. https://doi.org/10.1016/j.jprot.2018.09.011
Gupta S, Mishra SK, Misra S, Pandey V, Agrawal L, Nautiyal CS, Chauhan PS (2020) Revealing the complexity of protein abundance in chickpea root under drought-stress using a comparative proteomics approach. Plant Physiol Biochem 151:88–102. https://doi.org/10.1016/j.plaphy.2020.03.005
Steinfeld L, Vafaei A, Rösner J, Merzendorfer H (2019) Chitin prevalence and function in bacteria, fungi and protists. Adv Exp Med Biol 1142:19–59. https://doi.org/10.1007/978-981-13-7318-3_3
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115(2):327–334. https://doi.org/10.1104/pp.115.2.327
Wang Q, Guo Q, Guo Y, Yang J, Wang M, Duan X, Niu J, Liu S, Zhang J, Lu Y, Hou Z, Miao W, Wang X, Kong W, Xu X, Wu Y, Rui Q, La H (2018) Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OPEN STOMATA 1. J Exp Bot 69(18):4403–4417. https://doi.org/10.1093/jxb/ery205
Teale W, Palme K (2018) Naphthylphthalamic acid and the mechanism of polar auxin transport. J Exp Bot 69(2):303–312. https://doi.org/10.1093/jxb/erx323
Kidrič M, Sabotič J, Stevanović B (2014) Desiccation tolerance of the resurrection plant Ramonda serbica is associated with dehydration-dependent changes in levels of proteolytic activities. J Plant Physiol 171(12):998–1002. https://doi.org/10.1016/j.jplph.2014.03.011
Pichler G, Stöggl W, Candotto Carniel F, Muggia L, Ametrano CG, Holzinger A, Tretiach M, Kranner I (2020) Abundance and extracellular release of phytohormones in aero-terrestrial microalgae (Trebouxiophyceae, Chlorophyta) as a potential chemical signaling source. J Phycology (in press). https://doi.org/10.1111/jpy.13032
Hajheidari M, Eivazi A, Buchanan BB, Wong JH, Majidi I, Salekdeh GH (2007) Proteomics uncovers a role for redox in drought tolerance in wheat. J Proteome Res 6(4):1451–1460. https://doi.org/10.1021/pr060570j
Pinheiro C, Kehr J, Ricardo CP (2005) Effect of water stress on lupin stem protein analysed by two-dimensional gel electrophoresis. Planta 221(5):716–728. https://doi.org/10.1007/s00425-004-1478-0
Bray EA (2002) Classification of genes differentially expressed during water-deficit stress in Arabidopsis thaliana: an analysis using microarray and differential expression data. Ann Bot 89 Spec No:803-811. https://doi.org/10.1093/aob/mcf104
Schaller A (2004) A cut above the rest: the regulatory function of plant proteases. Planta 220(2):183–197. https://doi.org/10.1007/s00425-004-1407-2
Augusti A, Scartazza A, Navari-Izzo F, Sgherri CL, Stevanovic B, Brugnoli E (2001) Photosystem II photochemical efficiency, zeaxanthin and antioxidant contents in the poikilohydric Ramonda serbica during dehydration and rehydration. Photosynth Res 67(1-2):79–88. https://doi.org/10.1023/A:1010692632408
Sgherri C, Stevanovic B, Navari-Izzo F (2004) Role of phenolics in the antioxidative status of the resurrection plant Ramonda serbica during dehydration and rehydration. Physiol Plant 122(4):478–485. https://doi.org/10.1111/j.1399-3054.2004.00428.x
Beckett RP, Minibayeva FV, Vylegzhanina NN, Tolpysheva T (2003) High rates of extracellular superoxide production by lichens in the suborder Peltigerineae correlate with indices of high metabolic activity. Plant Cell Environ 26(11):1827–1837. https://doi.org/10.1046/j.1365-3040.2003.01099.x
Minibayeva F, Beckett RP (2001) High rates of extracellular superoxide production in bryophytes and lichens, and an oxidative burst in response to rehydration following desiccation. New Phytol 152(2):333–341. https://doi.org/10.1046/j.0028-646X.2001.00256.x
Jovanović Ž, Rakić T, Stevanović B, Radović S (2011) Characterization of oxidative and antioxidative events during dehydration and rehydration of resurrection plant Ramonda nathaliae. Plant Growth Regul 64(3):231–240. https://doi.org/10.1007/s10725-011-9563-4
Kärkönen A, Kuchitsu K (2015) Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 112:22–32. https://doi.org/10.1016/j.phytochem.2014.09.016
Vanden Wymelenberg A, Sabat G, Mozuch M, Kersten PJ, Cullen D, Blanchette RA (2006) Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol 72(7):4871–4877. https://doi.org/10.1128/AEM.00375-06
Davidson R, Reeves P, Manosalva P, Leach J (2009) Germins: a diverse protein family important for crop improvement. Plant Sci 177(6):499–510. https://doi.org/10.1016/j.plantsci.2009.08.012
Pei Y, Li X, Zhu Y, Ge X, Sun Y, Liu N, Jia Y, Li F, Hou Y (2019) GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium wilt pathogens. Front Plant Sci 10:583. https://doi.org/10.3389/fpls.2019.00583
Liszkay A, van der Zalm E, Schopfer P (2004) Production of reactive oxygen intermediates (O2˙−, H2O2, and ˙OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol 136(2):3114–3123. https://doi.org/10.1104/pp.104.044784
Müller K, Linkies A, Vreeburg RAM, Fry SC, Krieger-Liszkay A, Leubner-Metzger G (2009) In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol 150(4):1855–1865. https://doi.org/10.1104/pp.109.139204
Schopfer P (2001) Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: Implications for the control of elongation growth. Plant J 28:679–688. https://doi.org/10.1046/j.1365-313x.2001.01187.x
Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé JP, Fink GR, Foster KR, Verstrepen KJ (2008) FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cells 135:726–737. https://doi.org/10.1016/j.cell.2008.09.037
Cardon ZG, Peredo EL, Dohnalkova AC, Gershone HL, Bezanilla M (2018) A model suite of green algae within the Scenedesmaceae for investigating contrasting desiccation tolerance and morphology. J Cell Sci 131(7):jcs212233. https://doi.org/10.1242/jcs.212233
Domozych DS, Domozych CE (2014) Multicellularity in green algae: upsizing in a walled complex. Front Plant Sci 5:649. https://doi.org/10.3389/fpls.2014.00649
Alsufyani T, Califano G, Deicke M, Grueneberg J, Weiss A, Engelen AH, Kwantes M, Mohr JF, Ulrich JF, Wichard T (2020) Macroalgal-bacterial interactions: identification and role of thallusin in morphogenesis of the seaweed Ulva (Chlorophyta). J Exp Bot 71(11):3340–3349. https://doi.org/10.1093/jxb/eraa066
Boothby TC, Pielak GJ (2017) Intrinsically disordered proteins and DT: elucidating functional and mechanistic underpinnings of anhydrobiosis. Bioessays 39(11):10. https://doi.org/10.1002/bies.201700119
Koshland D, Tapia H (2019) Desiccation tolerance: an unusual window into stress biology. Mol Biol Cell 30(6):737–741. https://doi.org/10.1091/mbc.E17-04-0257
Schatz D, Rosenwasser S, Malitsky S, Wolf SG, Feldmesser E, Vardi A (2017) Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat Microbiol 2(11):1485–1492. https://doi.org/10.1038/s41564-017-0024-3
Rutter BD, Innes RW (2017) Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol 173(1):728–741. https://doi.org/10.1104/pp.16.01253
Acknowledgements
This study was supported by a grant from the Spanish Ministry of Science, Innovation and Universities [CGL2016-80259-P]. We thank the Proteomics Unit of Complutense University of Madrid, a member of ProteoRed (supported by Grant PRB3 [IPT17/0019 - ISCIII-SGEFI / ERDF]) for the LC-MS/MS analyses of EPS-associated proteins from TR9 and Csol algae, and Gabriel Casano for the English revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
González-Hourcade, M., del Campo, E.M. & Casano, L.M. The Under-explored Extracellular Proteome of Aero-Terrestrial Microalgae Provides Clues on Different Mechanisms of Desiccation Tolerance in Non-Model Organisms. Microb Ecol 81, 437–453 (2021). https://doi.org/10.1007/s00248-020-01604-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01604-8