Abstract
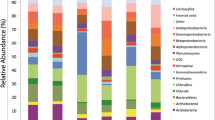
The Brazilian Cerrado is the second largest biome in Brazil and is considered a biodiversity hotspot. In this work, we compared the bacterial communities in Cerrado soil associated with four types of native vegetation (Cerrado Denso, Cerrado sensu stricto, Campo Sujo, and Mata de Galeria) by ribosomal RNA intergenic spacer analysis, terminal fragment restriction length polymorphism and pyrosequencing. The fingerprinting results were very similar. The bacterial communities of Cerrado Denso and Cerrado sensu stricto grouped together and were distinct from those in Campo Sujo and Mata de Galeria. Pyrosequencing generated approximately 40,000 16S rRNA gene sequences per sample and allowed the identification of 17 phyla in soil samples under Cerrado vegetation. Acidobacteria were dominant in all areas studied with a relative frequency of 40–47 %, followed closely by Proteobacteria accounting for 34–40 % of the sequences. Results from all molecular techniques used suggested that the bacterial communities of Cerrado sensu stricto and Cerrado Denso are very similar to each other, while Campo Sujo forms a separate group, and Mata de Galeria is the most distinct with higher species richness. This is the first extensive study of native Cerrado soil microbiota, an important but endangered biome.





Similar content being viewed by others
References
Coutinho LM (2006) O conceito de bioma. Acta bot bras 20(1):13–23
Cole MM (1986) The Savannas: biogeography and geobotany. Academic, London
Mistry J (2000) World savannas: ecology and human use. Longman (Pearson Education)-Prentice Hall, Harlow
Eiten G (1972) The Cerrado vegetation of Brazil. Bot Rev 38(2):201–341
Ribeiro JF, Walter BMT, Sano SMA (2008) Fitosionomias do Bioma Cerrado. In: Almeida SP (ed) Cerrado: ecologia e flora. Embrapa Cerrados, Planaltina, pp 89–166
Haridasan M (1994) Solos do Distrito Federal. In: Novaes-Pinto M (ed) Cerrado: Caracterização, ocupação e perspectivas - O caso do Distrito Federal, 2nd edn. Editora Universidade de Brasília/SEMATEC, Brasília, pp 321–344
Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12(11):2998–3006
Schloss PD, Handelsman J (2006) Toward a census of bacteria in soil. PLoS Comput Biol 2(7):e92–e92
Morales SE, Cosart TF, Johnson JV, Holben WE (2009) Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl Environ Microbiolol 75:668–675
Borneman J, Triplett EW (1997) Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol 63(7):2647–2653
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW 1 (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75(15):5111–5120
Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6(2):e17000
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16 S rRNA. Appl Environ Microbiol 63(11):4516–4522
Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci USA 103(32):12115–12120
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304(5667):66–74
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428(6978):37–43
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8(7):R143
Schloss P (2010) The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16 S rRNA gene-based studies. PLoS Comput Biol 6(7):1–16
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12(1):118–123
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT (2009) Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6(9):639–644
Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF (2004) Divergence and redundancy of 16 S rRNA sequences in genomes with multiple RRN OPERONs. J Bacteriol 186(9):2629–2635
Gutell RR, Larsen N, Woese CR (1994) Lessons from an evolving rRNA: 16 S and 23 S rRNA structures from a comparative perspective. Microbiol Rev 58(1):10–26
Ratter JA, Bridgewater S, Ribeiro JF (2003) Analysis of the floristic composition of the brazilian cerrado vegetation III: comparison of the woody vegetation of 376 areas. Edinb J Bot 60(1):57–109
Castro AAJF, Martins FR, Tamashiro JY, Shepherd GJ (1999) How rich is the flora of the Brazilian cerrados? Ann Mo Bot Gard 86:192–224
Briani D, Palma A, Vieira E, Henriques R (2004) Post-fire succession of small mammals in the Cerrado of central Brazil. Biodivers Conserv 13(5):1023–1037
Blamires D, de Oliveira G, de Souza BB, Diniz-Filho JAF (2008) Habitat use and deconstruction of richness patterns in Cerrado birds. Acta Oecologica 33(1):97–104
Castro AP, Quirino BF, Pappas G, Kurokawa AS, Neto EL, Krüger RH (2008) Diversity of soil fungal communities of Cerrado and its closely surrounding agriculture fields. Arch Microbiol 190:129–139
Quirino BF, Pappas GJ, Tagliaferro AC, Collevatti RG, Neto EL, da Silva MR, Bustamante MM, Kruger RH (2009) Molecular phylogenetic diversity of bacteria associated with soil of the savanna-like Cerrado vegetation. Microbiol Res 164:59–70
Bresolin JD, Bustamante MMC, Krüger RH, Silva MRSS, Perez KS (2010) Structure and composition of bacterial and fungal community in soil under soybean monoculture in the Brazilian Cerrado. Braz J Microbiol 41:391–403
Peixoto RS, Chaer GM, Franco N, Junior FBR, Mendes IC, Rosado AS (2010) A decade of land use contributes to changes in the chemistry, biochemistry and bacterial community structures of soils in the Cerrado. Anton Leeuw Int J G 98(3):403–413
Myers N, Mittermeier RA, Mittermeier CG, Fonseca GABd, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Grüntzig V, Stres B, del Rio HL Ayala, Tiedje JM (2002) Improved protocol for T-RFLP by capillary electrophoresis. Center for Microbial Ecology, Michigan State University, East Lansing, Michigan
Culman S, Bukowski R, Gauch H, Cadillo-Quiroz H, Buckley D (2009) T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10(1):171
Armougom F, Raoult D (2009) Exploring microbial diversity using 16 S rRNA high-throughput methods. J Comput Sci Syst Biol 2(1):74–92
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Blankenberg D, Kuster GV, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. Chap 19 (Unit 19.10): 1–21
Reeder J, Knight R (2010) Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7(9):668–669
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267
Magurran AE (2004) Measuring biological diversity. Blackwell Science, Oxford
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Parks DH, Beiko RG (2010) Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26(6):715–721
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16 S rRNA and 16 S rRNA genes. Appl Environ Microbiol 72(3):1719–1728
Bruce T, Martinez I, Maia Neto O, Vicente A, Kruger R, Thompson F (2010) Bacterial community diversity in the Brazilian Atlantic forest soils. Microb Ecol 60(4):840–849
Quaiser A, Ochsenreiter T, Lanz C, Schuster SC, Treusch AH, Eck J, Schleper C (2003) Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol Microbiol 50(2):563–575
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3(4):442–453
Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, Barabote RD, Bradley B, Brettin TS, Brinkac LM, Bruce D, Creasy T, Daugherty SC, Davidsen TM, DeBoy RT, Detter JC, Dodson RJ, Durkin AS, Ganapathy A, Gwinn-Giglio M, Han CS, Khouri H, Kiss H, Kothari SP, Madupu R, Nelson KE, Nelson WC, Paulsen I, Penn K, Ren Q, Rosovitz MJ, Selengut JD, Shrivastava S, Sullivan SA, Tapia R, Thompson LS, Watkins KL, Yang Q, Yu C, Zafar N, Zhou L, Kuske CR (2009) Three genomes from the phylum acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75(7):2046–2056
Sait M, Davis KE, Janssen PH (2006) Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl Environ Microbiol 72(3):1852–1857
Zhang L, Xu Z (2008) Assessing bacterial diversity in soil. J Soils Sediments 8(6):379–388
Martinelli LA, Howarth RW, Bustamante MMC, Medina E, Asner GP, Nardoto GB, Garcia-Montiel DC (2006) Nitrogen cycling in tropical and temperate savannas. In: Nitrogen cycling in the Americas: natural and anthropogenic influences and controls. Springer, Dordrecht, pp. 209–237
Nardoto GB, da Cunha Bustamante MM, Pinto AS, Klink CA (2006) Nutrient use efficiency at ecosystem and species level in savanna areas of Central Brazil and impacts of fire. J Trop Ecol 22(02):191–201
Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, Dv S (2007) Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71(3):495–548
Holmes AJ, Bowyer J, Holley MP, O’Donoghue M, Montgomery M, Gillings MR (2000) Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol Ecol 33(2):111–120
Gremion F, Chatzinotas A, Harms H (2003) Comparative 16 S rDNA and 16 S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ Microbiol 5(10):896–907
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67(10):4399–4406
Haridasan M (1998) Solos de mata de galeria e nutrição mineral de espécies arbóreas em condições naturais. In: Ribeiro JF (ed) Cerrado: matas de galeria. EMBRAPA-CPAC, Planaltina, pp 19–28
Felfili JM (1995) Diversity, structure and dynamics of a gallery forest in central Brazil. Vegetatio 117:1–15
Resende J, Markewitz D, Klink C, Bustamante M, Davidson E (2010) Phosphorus cycling in a small watershed in the Brazilian Cerrado: impacts of frequent burning. Biogeochemistry 105:105–118
Parron LM (2004) Aspectos da ciclagem de nutrientes em função do gradiente topográfico, em uma Mata de galeria no Distrito Federal. Universidade de Brasília, Brasília
Singleton DR, Furlong MA, Peacock AD, White DC, Coleman DC, Whitman WB (2003) Solirubrobacter pauli gen. nov., sp. nov., a mesophilic bacterium within the Rubrobacteridae related to common soil clones. Int J Syst Evol Microbiol 53:485–490
Guerrero G, Peralta H, Aguilar A, Diaz R, Villalobos M, Medrano-Soto A, Mora J (2005) Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales. BMC Evol Biol 5(1):55
Hanada S, Takaichi S, Matsuura K, Nakamura K (2002) Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol 52:187–193
Garbeva P, van Veen JA, van Elsas JD (2004) Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270
Singh BK, Munro S, Potts JM, Millard P (2007) Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl Soil Ecol 36:147–155
Acknowledgments
This work was supported by grants from FAPDF/CNPq. JFA acknowledges a fellowship from Ouro Fino Agronegócio. APC acknowledges a fellowship from CAPES. We acknowledge Dr. Ashley Shade for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(DOC 51 kb)
Table S2
(DOC 98 kb)
Figure S 1
Structure of Cerrado vegetation physiognomies. a Pictures of areas. b Illustration of vegetation cover and topography of study areas (PDF 600 kb)

Figure S 2
Principal component analysis of soil physicochemical proprieties of Cerrado vegetation physiognomies. (CS1 Campo Sujo “1 to 5,” CS2 Campo Sujo “6 to 10,” CD1 Cerrado Denso “1 to 5,” CD2 Cerrado Denso “6 to 10,” SS1 Cerrado sensu stricto “1 to 5,” SS2 Cerrado sensu stricto “6 to 10,” MG1 Mata de Galeria “1 to 5,” and MG2 Mata de Galeria “6 to 10”) (JPEG 1905 kb)

Figure S3
Relative frequency of Acidobacteria subdivisions in the soil of different Cerrado vegetation physiognomies based on pyrosequencing data. The classifier of Ribosomal Database Project was used to assign sequences to the different subdivisions. Each Cerrado type was assigned a different color, and the replicate samples are indicated by different shades of the same color. The length of each color segment represents the percentage of that particular Acidobacteria subdivision found in each Cerrado type. Above each column, the total number of sequences identified for each Acidobacteria subdivision is shown (n) (JPEG 2033 kb)
Figure S4
Significant differences between the relative frequencies of bacterial orders of a comparison of each vegetation physiognomy by STAMP software. a Mata de Galeria and Campo Sujo. b Mata de Galeria and Cerrado sensu stricto. c Mata de Galeria and Cerrado Denso. d Campo Sujo and Cerrado sensu stricto. e Campo Sujo and Cerrado Denso. f Cerrado sensu stricto and Cerrado Denso (PDF 1477 kb)
Rights and permissions
About this article
Cite this article
Araujo, J.F., de Castro, A.P., Costa, M.M.C. et al. Characterization of Soil Bacterial Assemblies in Brazilian Savanna-Like Vegetation Reveals Acidobacteria Dominance. Microb Ecol 64, 760–770 (2012). https://doi.org/10.1007/s00248-012-0057-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0057-3