Abstract
Here we analyze the molecular evolution of the β-esterase gene cluster in the Drosophila genus using the recently released genome sequences of 12 Drosophila species. Molecular evolution in this small cluster is noteworthy because it contains contrasting examples of the types and stages of loss of gene function. Specifically, missing orthologs, pseudogenes, and null alleles are all inferred. Phylogenetic analyses also suggest a minimum of 9 gene gain–loss events; however, the exact number and age of these events is confounded by interparalog recombination. A previous enigma, in which allozyme loci were mapped to β-esterase genes that lacked catalytically essential amino acids, was resolved through the identification of neighbouring genes that contain the canonical catalytic residues and thus presumably encode the mapped allozymes. The originally identified genes are evolving with selective constraint, suggesting that they have a “noncatalytic” function. Curiously, 3 of the 4 paralogous β-esterase genes in the D. ananassae genome sequence have single inactivating (frame-shift or nonsense) mutations. To determine whether these putatively inactivating mutations were fixed, we sequenced other D. ananassae alleles of these four loci. We did not find any of the 3 inactivating mutations of the sequenced strain in 12 other strains; however, other inactivating mutations were observed in the same 3 genes. This is reminiscent of the high frequency of null alleles observed in one of the β-esterase genes (Est7/EstP) of D. melanogaster.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The tightly linked esterase 6 (Est6) and esterase 7 (Est7) genes, which comprise the β-esterase cluster of Drosophila melanogaster and its sibling species, have each been intensively studied as models of microevolutionary change for different reasons. Est6 has acquired a novel function in the melanogaster species subgroup (Oakeshott et al. 2000), where it is highly expressed in the sperm ejaculatory duct of the adult male fly. The encoded enzyme is transferred to the female fly during mating and modifies her subsequent egg-laying and remating behaviors (Meikle et al. 1990; Richmond et al. 1990; Saad et al. 1994). Est7 is predominantly expressed in integumental tissue of late larvae and early pupae (Dumancic et al. 1997). Its function there is unknown; however, there are high frequencies of Est7 null alleles (>60% in one D. melanogaster population) containing a range of apparently disabling mutations (Balakirev and Ayala 1996). These data might suggest that the function of the EST7 protein has become redundant and that the gene is decaying toward pseudogene status (Balakirev and Ayala 2003; Balakirev et al. 2006). An alternative explanation suggested by Balakirev and Ayala (2004) is that Est7 maintains a function that is not disabled by these premature stop codons or frame-shifting mutations.
Apart from their respective gain and loss of function in the melanogaster subgroup, Est6 and Est7 have been essentially conserved as a tandem duplication through the rest of the melanogaster species group (Oakeshott et al. 1995, 1999; Balakirev et al. 2006). However, biochemical and molecular data indicate that the cluster is more elaborate in both composition and function elsewhere in the genus. Molecular data for D. pseudoobscura (obscura group, also in the subgenus Sophophora) shows a tandem triplication, with evidence of gene conversion or reciprocal recombination among paralogs (Brady and Richmond 1992; King 1998). One member encodes the major adult haemolymph β-esterase (as per Est6 in other subgroups of the melanogaster group); however, no data exist regarding the functions of the other two (Brady and Richmond 1990; Tamarina et al. 1997). Isozyme data for D. virilis (virilis group) and several cactophilic species (repleta group) of the subgenus Drosophila suggest that many of these species have both an adult haemolymph and a preadult integument β-esterase, plus at least one and possibly as many as three β-esterases in the male reproductive tract, albeit these are found in the ejaculatory bulb rather than ejaculatory duct as in D. melanogaster (Oakeshott et al. 1990, 1993). It therefore appears that male fly reproductive tract activities have evolved more than once among the members of the β-esterase cluster in the genus.
Most remarkably, however, there is evidence that the cluster also includes members with novel, noncatalytic functions in the subgenus Drosophila. Molecular work to date on D. virilis and the cactophilic D. buzzatii has only recovered β-esterase genes, whose products lack the key “catalytic triad” residues that are required for esterase activity. In the case of D. virilis, the cloned gene is located at the same chromosome region as the one to which classical genetics had mapped the gene encoding the major ejaculatory bulb β-esterase isozyme; the inactive gene is likewise highly expressed in this tissue (Enikolopov et al. 1989; Sergeev et al. 1993, 1995). The data for D. buzzatii showed two tandemly arranged β-esterase genes, both lacking functional catalytic triads (East et al. 1990). However, the D. buzzatii genes, like the D. virilis one (and unlike the Est7 null alleles in D. melanogaster; Balakirev and Ayala 1996), still show attributes of genes under functional constraint (e.g., substitutions per synonymous site occur at a greater rate than those per nonsynonymous site).
Given these intriguing hints of diverse evolutionary trajectories for Drosophila β-esterases, we have identified and annotated the β-esterase sequences in the 12 species genome data set (Clark et al. 2007; Stark et al. 2007), compared them with the β-esterase sequences in the literature, and reconstructed a phylogeny of the β-esterases across the genus. This analysis leads us to 3further questions: (1) Have there been as many gene gain and loss events as the shortest gene tree suggests, or does reticulate evolution (such as gene conversion) play a role in homogenizing paralogs within species? (2) Given that the noncatalytic members all fall into one clade, what is the evidence that they do have a protein coding function? (3) Are the β-esterase genes that contain premature stop codons and frame-shifts in the genome sequence data pseudogenes or null alleles?
Materials and Methods
Sequence Analysis
BlastP was used with D. melanogaster EST6 (NP_788500) as a query to identify complete β-esterase sequences in the NCBI databases, and the following sequences were extracted: D. mauritiana (L10671, AY695921), D. teissieri (AY695922), D. orena (AY695924), D.affinis (AY754521), and D. miranda (AF016108, AY754520, AF016109). The sequences from D. buzzatii were obtained from East et al. (1990). D. melanogaster Est6, D. melanogaster Est7, and D. virilis EstS (Q05487) were then used as queries in tblastn searches of the CAF1 release of the Whole Genome Shotgun of 12 Drosophila genomes (http://www.flybase.org/blast/). Contigs with high similarity (generally blast scores >500) were downloaded, and the esterase sequences and neighboring genes were then annotated using Artemis version 7 (Rutherford et al. 2000). BlastP of the inferred amino acid sequences against the D. melanogaster protein databases confirmed the “β-esterase” classification. The table of blast hits generally ranked sequences orthologous to D. melanogaster CG8424 (JHEdup, a paralog to the β-esterases; Campbell et al. 2001) as the next highest hits.
The above-mentioned amino acid sequences were aligned with the four Anopheles gambiae esterases closest to the Drosophila β-esterases (EAU76436, EAL39807, EAA11835, and EAL39808; Ranson et al. 2002), the closest esterase from Aedes aegypti (EAT35318), the two Apis mellifera β-esterases (GB15327, GB10820; Claudianos et al. 2006), and D. melanogaster CG8424 (JHEdup) and CG6414 (which occurs in a clade with β-esterases and pheromone esterases in the analysis of Claudianos et al. 2006) using ClustalX (Larkin et al. 2007). Phylogenetic analysis demonstrated that mosquito and bee sequences are outgroups to the Drosophila β-esterase radiation (date not shown). Because the outgroup sequences (bees, mosquitoes, CG6414, and CG8424) introduced multiple indels, all were excluded from subsequent analysis except for one Anopheles sequence (EAU76436), and the remaining sequences were realigned. The sequences were trimmed at both ends, where indels made homologous site alignment uncertain, and the remaining gaps were handled as missing characters (thus, only sites aligning to 24-DDLP . . . IDGC-535 of D. melanogaster EST6 were included). Bayesian trees were constructed using MrBayes (Ronquist and Huelsenbeck 2003) with previous probabilities set so that substitution rates were fixed, but the state frequencies were allowed to vary (“Equalin” model). The model was set so that across-sites rates followed a gamma distribution with some invariant sites (rates = invgamma). Parsimony and neighbor-joining trees were constructed using PAUP* (Swofford 2003). Maximum likelihood trees were constructed using protML from Phylip 3.66 (Felsenstein 1989) using a Jones–Taylor–Thornton probability model and setting the across-sites rates to follow a gamma distribution with some invariant sites.
Sliding window plots (with window size of 7 amino acids) were constructed from amino acid alignments using Geneious Pro 3.8.5 (http://www.geneious.com). All of the Drosophila β-esterases were aligned, and sites corresponding to deletions in the noncatalytic esterases were removed. Two plots were generated, one with all five noncatalytic esterases (D. buzzatii and D. mojavensis ESTA and ESTB, and D. virilis ESTS) and another with all the other Drosophila β-esterases.
Mrtrans (Pearson 1990) was used to align nucleotide sequences according to the amino acid sequence alignment. dN and dS were calculated using the Nei and Gojobori method as implemented in PAML (Yang and Nielsen 2002). Trees based on the nucleotide sequences were created using neighbor-joining, parsimony, and maximum likelihood approaches. The appropriate model for the maximum likelihood tree was determined using Modeltest (Posada and Crandall 1998), which suggested that across-sites rates followed a gamma distribution, with some invariant sites following the general time-reversible model (GTR + I+G).
The minimum number of gene gain–loss events was also calculated based on the number of β-esterase genes observed in each species genome and the unrooted species tree, i.e., (((melanogaster subgroup = 2 genes, D. ananassae = 4 genes), obscura group = 3 genes), D. willistoni = 1 gene), ((D. mojavensis = 6 genes, D. virilis = 5 genes), D. grimshawi = 1 gene). The possible number of genes existing at each of the five ancestral nodes on the species tree was then considered (e.g., the ancestor of the melanogaster subgroup species and D. ananassae could have had two, three, or four genes, etc.). We then calculated which of these reconstructions (we explicitly enumerated 118 scenarios) invoked the fewest number of gene gain–loss events. We assumed gain and loss occurred at equal likelihood.
Analysis of Reticulate Evolution
The nucleotide alignment of the β-esterases was imported into Splits tree v4.1 (http://www.splitstree.org; Huson and Bryant 2006). Specific taxa were filtered out, and the pairwise homology index (PHI) test performed (Bruen et al. 2006). The distribution of splits across the D. ananassae alignment was obtained from the “show mutations” feature within the “recombination network” analysis and then imported into Excel for graphing. In this graph, the variable sites in the alignment of the four sequences are classified into seven types or “splits.” One split is when gene 1 differs from gene 2, gene 3, and gene 4 at a site. There is a split for each sequence (i.e., the four singleton splits). Then there is a split that groups gene 1 with gene 2, another that groups gene 1 with gene 3, and a third that groups gene 1 with gene 4. The graph allows the distribution of phylogenetic signal across the alignment to be observed. We also used the Recombination Detection Program (http://darwin.uvigo.es/rdp/rdp.html; Martin et al. 2005) to confirm the inferred recombination events.
Allele Sequencing
The D. ananassae lines used for this analysis included nine stock centre lines that had been collected from locations in the Americas, Asia. and Africa between 1966 and 2006, along with three Australian lines collected by C. R.’s laboratory in 2006 (Supplementary Table 1). DNA was extracted from single-fly homogenates after incubation with Chelex-100 (Bio-Rad, Hercules, CA). Briefly, 150 μL chelex solution was added to flies that had been crushed in 6 μL 3 mg/mL proteinase K. After incubation at 55°C for 1 hour, a brief vortex, and incubation at 95°C for 10 minutes, the homogenate was centrifuged for 3 minutes at 16 rcf and the supernatant, containing the genomic DNA, was extracted. Primers listed in Supplementary Table 2 were used to amplify approximately1.2 kb of each of the four esterase genes. Extra primers (Supplementary Table 2) were used to amplify the remainder of the Est6c gene. Polymerase chain reactions (PCRs) were carried out in volumes of 50 μL, with a final concentration of 1 × NEB ThermoPol PCR buffer; 5 U Taq polymerase, 200 μM each dNTP; and 0.2 M each primer. Cycling conditions were as follows: 94°C for 2 minutes; 35 cycles at 94°C for 15 seconds, 58°C for 15 seconds, and 72°C for 1 minute. The products were sent to Macrogen (Seoul, Korea) for sequencing from both ends. Sequences that contained indels were cloned into JM109 (Promega) competent cells using the pGEM-T easy kit (Promega) and resequenced. Sequences were assembled and aligned using Sequencher version 4.6 and MEGA4 (Tamura et al. 2007; http://www.megasoftware.net/) before being imported into DNAsp 4.10 (Rozas et al. 2003) for the calculation of population genetic statistics.
Results and Discussion
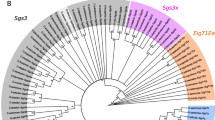
The β-Esterases of the Drosophila Genus
To reconstruct the evolution of the β-esterases within the Drosophila genus, we aligned Drosophila β-esterases previously described in the literature (East et al. 1990; Balakirev et al. 2005; Claudianos et al. 2006), orthologous esterases from mosquitoes and bees (Ranson et al. 2002; Claudianos et al. 2006), and sequences that we identified and annotated in the genome sequences of the 12 Drosophila species. The number of β-esterases we identified in Drosophila genomes ranges from one (D. willistoni and D. grimshawi) to six (D. mojavensis; Fig. 1). Paralogs within any one genome are found in a single tandem cluster, with tail-to-head orientation. Each of the Drosophila β-esterase genes has a single phase 1 intron, located at a single homologous site. This intron site is not found in other Drosophila esterases or the closest Anopheles esterases; however, is one of seven introns in the closest Apis esterases. Microsynteny supports the orthologous relation of these clusters within the Sophophoran subgenus; the clusters are flanked by CG6910 and CG5632 in all nine Sophophorans examined. However, the genes flanking the β-esterases in D. grimshawi (a Hawaiian Drosophila not yet assigned to a subgenus) and the two Drosophila subgenus species (D. mojavensis and D. virilis) are different (CG33173 and CG4702), suggesting that rearrangements occurred after the divergence of the two subgenera.
The β-esterase gene cluster in various Drosophila species. Arrows representing genes are pointing in the direction of transcription, and the β-esterase genes are labeled with the gene name. Genes flanking the β-esterase gene cluster are shaded to indicate orthology. The lines on which the arrows are drawn represent contiguous sequence. The dotted arrow in D. grimshawi represents a decayed pseudogene. Crosses (X) represent genes that appear to have inactivating mutations. The slash symbol indicates the β-esterase genes that appear to lack at least one catalytic triad residue necessary for esterase activity
The sequences of the β-esterases in the melanogaster and obscura groups of the subgenus Sophophora have been well characterized (Collet et al. 1990; Karotam et al. 1993; Balakirev et al. 2005). In the Drosophila subgenus, one D. virilis esterase has been fully characterized (ESTS; Enikolopov et al. 1989; Sergeev et al. 1995); partial genomic sequences are available for two D. buzzatii β-esterase genes (EstA and EstB; East et al. 1990); and partial amino acid sequences have been obtained for EST4 and EST5 of D. mojavensis (Pen et al. 1986). Because the genomes of D. virilis and D. mojavensis are among the 12 Drosophila species genomes sequenced, we can confirm, re-annotate, and complete the full coding sequences of these genes.
The closest match to EstS among the 5 D. virilis esterases of the β-esterase cluster annotated here is that adjacent to the CG33173 ortholog (to the far left of the cluster in Fig. 1). The ESTS amino acid sequence differs from that encoded by this gene at 27 amino acid sites. However, 9 of these are the N-terminal amino acids, and 11 are clustered at a single site and are consistent with frame-shift errors introduced during the original sequencing of EstS. We therefore believe that the gene closest to CG33173 in D. virilis genomic sequence is allelic to the EstS gene (Supplementary Fig. 1).
Pen et al. (1986) determined 38 residues from D. mojavensis Esterase 5 (EST5) by Edman degradation, although identification of 7 of these was “tentative.” The best match for this in the D. mojavensis genome is a match of 33 (the next best is 28), where 4 of the mismatches occur in “tentatively” assigned peptide residues, and the fifth occurs at a site that is either phenylalanine or tyrosine in all β-esterases, except in the peptide sequences of Pen et al. (1986), which have the site as an alanine. Therefore, EST5 is most likely encoded by Est2a (Supplementary Fig. 2).
Pen et al. (1986) also obtained sequence for D. mojavensis esterase 4 (EST4); however, identifying the sequence encoding it in the genome is less clear because there are two translated genomic sequences (EST2B and EST2C) that each differ from the peptide sequence at two sites confidently ascertained by Pen (Supplementary Fig. 2). However, the empirically determined amino acid composition of EST4 matches the composition of translated Est2c better than that of Est2b and leads us to tentatively suggest that EST4 is encoded by Est2c (Supplementary Table 3).
Three β-esterases have been previously described as lacking amino acids vital for catalytic function. ESTS from D. virilis and ESTB from D. buzzatii lack the catalytic triad histidine; ESTB also lacks the catalytic triad acid; and D. buzzatii ESTA and ESTB do not have the catalytic triad serine. The recent sequencing of the genomes of D. virilis and D. mojavensis confirms the existence of esterases that lack these residues key to catalysis. In addition, the sequence that we believe to be allelic to ESTS also has glycine at the site where the catalytic serine is usually found (Supplementary Fig. 3).
The Phylogeny of the β-Esterases
Figure 2 shows a phylogeny of the β-esterase genes generated from amino acid sequences using a Bayesian approach. There are five major clades in this reconstruction: EST6 of the melanogaster group, EST7 of the melanogaster group, EST5 of the obscura group, EST2 of the Drosophila subgenus (which also includes D. grimshawi), and the EST1 clade of the Drosophila subgenus. Within the EST1 clade, there is also a subclade containing the five proteins that lack the catalytic triad residues. Notwithstanding this clear and simple overall topology, the tree also shows many changes in the β-esterase gene complements through the course of drosophilid evolution. Thus, the tree suggests that there have been five gene duplications in the Sophophoran subgenus and six in the subgenus Drosophila. The five in the Sophophoran subgenus include two in the obscura group leading to Est5a/5b/5c and one leading to Est6 and Est7 and two in the D. ananassae lineage to give Est6a/6b/6c. The six gene duplications in the Drosophila subgenus include one forming the EST1 and EST2 clades, two in the EST1 clade, and three in the EST2 clade. The noncatalytic esterases from the Drosophila subgenus form a single clade spawned from the duplication of an ancestral Est1.
A Bayesian phylogeny of the β-esterases of the Drosophila genus. All branches with clade credibility (posterior probability) values < 70 are collapsed. The exact clade credibility scores are shown for nodes inferred to represent gene-duplication events (circles) and at the branch representing the origin of the noncatalytic clade (diamond). The asterisk represents the point at which the Anopheles gambiae outgroup joins the tree in the maximum likelihood analysis we performed (data not shown)
This phylogeny also suggests that there has been a loss of an Est1-like gene in the D. grimshawi lineage. This is because the Est1/Est2 duplication predates the divergence of D. grimshawi from the Drosophila subgenus; however, D. grimshawi has only a single active β-esterase gene that is in the EST2 clade. There is a highly degenerated pseudogene upstream of this gene, but the pseudogene is most similar to this active D. grimshawi Est2 gene and so appears not to be a relic of Est1.
Various other methods (maximum likelihood, maximum parsimony, and distance) were also used to infer phylogenetic relations of the β-esterases. All methods generally agree, and most branches are associated with high clade-confidence measures (posterior probability and bootstrap values); the gene trees mostly recapitulate the known species trees (these analyses are not shown). However, there are three differences in the topologies of the trees built with the different methods.
The first difference relates to the topology of the EST2 clade, where either three or four gene duplications are inferred. In all cases, D. mojavensis EST2A and D. mojavensis EST2B are most closely related to each other, and D. grimshawi has the basal position in the clade; however, the rest of the topology of this clade varies with the tree-building method. Compared with the three gene duplications inferred in Fig. 2, maximum parsimony analysis indicates that two duplications independently occurred in the D. mojavensis lineage and the D. virilis lineage to give Est2a, Est2b, and Est2c (D. mojavensis) and Est2d, Est2e, and Est2f (D. virilis).
The second way in which trees differ in their topology is the point at which the outgroup sequence joins the rest of the tree (Fig. 2). In maximum likelihood analysis, the outgroup divides the branch separating the EST1/noncatalytic clade from the rest of the tree. This would mean that the ancestral Drosophila would have had two esterase genes, one of which would have been lost in the Sophophoran subgenus (Fig. 2). In Bayesian and other analyses, the topology of the tree is consistent, with the ancestral Drosophila having a single esterase gene.
The final difference in tree topology between different methods is that in Bayesian analysis D. affinis is an outgroup to the EST5B and EST5C sequences rather than an ingroup of the EST5B sequences. A complete characterization of the D. affinis β-esterase cluster may resolve this minor discrepancy.
In summary, our reconciliation of the β-esterase gene tree with the species tree suggests 11 or 12 gene duplications and at least 1 gene loss (not including the D. grimshawi pseudogene, which may never have been functional). Our findings contrast with the evolutionary model proposed by Brady and Richmond (1992). In all of our analyses, the Est6/7 duplication occurred after the melanogaster/obscura group divergence, whereas Brady and Richmond have it preceding the divergence. The model of Brady and Richmond (1992) was based on the assertion that independent duplications in the melanogaster and obscura radiations were unlikely and that the life stage expression of Est7 and Est5a were both third-instar larvae. Therefore, they were orthologs. Similarly EST6 and EST5B, were the major β-esterase isozymes in their respective species and therefore orthologs. Importantly, Brady and Richmond (1992) also presented an argument of interparalog recombination between Est5a and Est5b.
Interparalog Recombination?
If intergene conversion or recombination does occur, then phylogenetic methods ignoring these process may produce a misleading history of gene gain and loss, making duplications appear younger than they actually are. Although our most parsimonious protein tree (Fig. 2) suggests that the Est6 and Est7 duplication occurs after the melanogaster/obscura divergence, a splits network based on the nucleotide sequences shows that there is also signal in the sequences supporting Brady and Richmond’s model, which groups EST5A and EST7 (Fig. 3). The proposition of interparalog recombination implied in this graphical representation is supported by a highly significant result in the PHI test of recombination, which examines the patterns of incompatibility between closely linked parsimony informative sites (p < 1.4 × 10−9; Bruen et al. 2006). Essentially, this result indicates that nearby sites (within 100 bp of each other) have greater genealogical correlations than distant sites. Although we expect to see evidence of parallel and backward mutations in data sets exhibiting as much divergence as this, the clustering of sites with the same genealogy supports the possibility of recombination between these genes.
We extended this analysis to test for reticulate evolution among paralogs of other species. Specifically, we used the PHI test (Bruen et al. 2006) to look for reticulate evolution in each of the three β-esterase gene clusters (D. ananassae, D. mojavensis, and D. virilis), each of which contain at least four genes (because four taxa are required to define “parsimony informative” sites). In each case, evidence for reticulate evolution (presumably gene conversion or intergenic recombination) was identified (D. ananassae p < 0.002, D. mojavensis p = 0.0, and D. virilis p < 0.009). Further analysis using a suite of other recombination detection programs that detect recombination through different methods (http://darwin.uvigo.es/rdp/rdp.html; Martin et al. 2005) support the argument for interparalog recombination and identify putative conversion tracts. Some of the putative conversion tracts were as short as 9 bp and were therefore unconvincing.
The most convincing case of an interparalog conversion event (i.e., the longest and most homogeneous conversion tract) occurs in the D. ananassae cluster (the average Bonferroni-corrected p values for the tests were RDP p = 1.2 × 10−4; Geneconv p = 1.4 × 10−4; and MaxiChi p = 3.37 × 10−3). The distribution of phylogenetic splits among the four D. ananassae sequences shows that between positions 120 and 420, Est6a and Est6b have a paucity of unique sites and an excess of sites that group them together (Fig. 4). This is consistent with a directional gene conversion where part of Est6a overwrote Est6b or vice versa. If this region is excluded from the PHI test, the p value is no longer significant (p = 0.55). Throughout the rest of the alignment support for competing trees is approximately equal and interdigitated. Thus, there is a large amount of homoplasy in the data, and at least some of it (i.e., a tract between 120 and 420) may be due to intergene exchange.
Distribution of the sites concurring with the seven possible splits in the alignment of four D. ananassae sequences are shown on separate rows. The four trivial splits (where a single sequence differs from the other three at a site) are shown as plus signs. The three alternate splits (which are incompatible with each other) are shown as triangles. The box indicates the site of a proposed intergene recombination event
An alternate explanation for such a pattern is that there is extreme variation in selective constraint across the gene. To illustrate with the D. ananassae genes, the region between 120 and 420 could have much greater selective constraint and may reflect the true genealogy, whereas the rest of the gene may have changed so much that homoplasy masks the true genealogical history. Such a pattern could result in a significant test with the PHI test. Although we cannot rule out this possibility, we note that the constraint must apply to the silent sites as well.
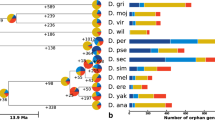
Thus, although there is some evidence for interparalog recombination, that signal is not clear enough (possibly because of age, length of recombination tracts, and/or variation in selective constraint) to factor such events into sequence-based phylogenies. However, the observed gene numbers in the Drosophila species alone suggests a minimum number of gene gain–loss events (i.e., independent of the sequences that compose them). If we assume an unrooted phylogeny of the Drosophila species (i.e., [[[melanogaster subgroup, ananassae], obscura group], wilistoni] [[mojavensis,virilis],grimshawi]]) and weight gene duplications and loss events equally, then a minimum of 9 gain/loss events are required to explain the observed gene composition of the β-esterases among the 12 Drosophila species. There are 5 scenarios in which the 9 changes can be reconstructed, including 1 with no losses, and 1 scenario with 4 gains and 5 losses (Supplementary Table 4). These scenarios differ by whether 1, 2, or 3 genes are inferred to be in the ancestral Drosophila. Some are also more likely than others once we consider the sequences again. For instance, 3 of the scenarios invoke a gene loss in the ancestor of the melanogaster subgroup after the ananassae divergence. If this has happened, we might expect to find some relic of a pseudogene in the genomes of the 5 melanogaster subgroup genomes. Thus, at a bare minimum, there have been 9 gain–loss events in the Drosophila radiation.
Functional Noncatalytic Homologs
Of all the β-esterases characterized, 5 are noncanonical at the catalytic triad sites (Supplementary Fig. 3) and, as shown in Fig. 2, they form a clade, suggesting a single origin before the divergence of the virilis and the repleta groups (D. buzzatii and D. mojavensis). We know that the D. virilis gene is transcribed because there are 16 Expressed Sequence Tags corresponding to it in the National Center for Biotechnology Information’s dbEST (in December 2008; e.g., EB569648). Although the proteins they encode are incapable of triad-mediated catalysis, the relative rates of nonsynonymous and synonymous divergence of these genes since the catalytic triad mutations indicate that they do have a protein-coding function (e.g., dN/dS for D. mojavensis EstA to D. mojavensis EstB is 0.27; D. virilis EstS to D. mojavensis EstA is 0.14; and D.virilis EstS to D. mojavensis EstB is 0.10). Furthermore, they are all approximately the same length as catalytic esterases, and the pattern of amino acid divergence within the clade exhibits selective constraint typical of a carboxylesterases, suggesting that they maintain core structural features of this protein family (Fig. 5).
Similarity plots showing that the divergence among the noncatalytic β-esterases correlates with the divergence profile of catalytic β-esterases. The x-axis represents position in the amino acid alignment of β-esterases when all gaps are removed. The y-axis records the level of amino acid similarity calculated with a seven–amino acid sliding window
There are in fact several precedents for the evolution of noncatalytic functions in other clades of the carboxyl/cholinesterase family to which the β-esterases belong (Claudianos et al. 2006). Most of these have ancient origins, predating the divergence of vertebrates and invertebrates, and most involve binding of protein ligands and functions in cell–cell adhesion in the developing nervous system (Gilbert and Auld 2005). However, there are also a few more recent precedents, most notably the glutactins of the higher Diptera, although functional information on these is largely lacking (Grisaru et al. 1999; Olson et al. 1990). Thus, this clade of noncatalytic β-esterases in the Drosophila subgenus represents an evolutionarily more recent acquisition of a presumably novel noncatalytic function.
D. ananassae Null Alleles or Pseudogenes?
Apart from the noncatalytic clade, all of the β-esterase sequences have the canonical active site residues, and most appear functional. However, three esterase genes have single mutations that disrupt their open reading frames and, remarkably, they are all in the D. ananassae gene cluster: Est6a has a 2nt insertion; Est6c has a stop codon; and Est7 has a 2nt deletion. Est6c also has a 6-codon deletion adjacent to a string encoding an unusual stretch of amino acids, 87-TAINT-91, that makes it aberrant relative to other esterases. All of these inactivating mutations are represented by multiple high-quality sequence traces. Thus, only one of the four β-esterase genes of the sequenced D. ananassae strain, namely Est6b, can encode a full-length functional esterase.
The three D. ananassae Est6 paralogs form a clade. However, the gene duplications are not recent because they are significantly diverged; the silent site divergence between the three D. ananassae Est6 paralogs range from 0.75 to 1 substitution/site. In comparison, the average divergence at four-fold degenerate sites in genome-wide comparisons of D. melanogaster with D. yakuba and D. ananassae are approximately 0.2 (diverged 6 to 15 mya; Lachaise et al. 1988) and approximately 1, respectively (Stark et al. 2007). Thus, it appears that the D. ananassae duplications happened many millions of years ago, possibly shortly after the D. ananassae and D. melanogaster lineages diverged. Furthermore, the genes seem to have been evolving with purifying selection because dN/dS for each pair wise comparison between the four paralogs is only 0.10 to 0.16, well below the value of 1 expected under neutrality.
To explore the possibility that the inactivating mutations in the D. ananassae genes may be polymorphic and represent null alleles within the stock that had its genome sequenced (stock number 14024–0371.13, which is referred to later as Matsuda AABBg1), we sequenced ≥ 1 kb more from each of Est6a, Est6b, Est6c, and Est7 in 12 D. ananassae lines from disparate global populations (Supplementary Table 2; GenBank accession numbers FJ884695 through FJ884743) to yield 16 allelic sequences for Est6a, 16 for Est6b, 14 for Est6c, and 16 for Est7. The inactivating mutations in the Matsuda AABBg1 line were not present in any of the other lines sequenced. However Est6a, Est6c, and Est7 each had alleles with other inactivating mutations among the 12 additional lines (see Fig. 6). No inactivating mutations could be found in Est6b in any of the lines sequenced. In this article, we refer to alleles harbouring polymorphic inactivating mutations as “null alleles,” unless there is also a fixed inactivating mutation, in which case we refer to the locus as a “pseudogene.”
Inactivating mutations found in the D. ananassae β-esterase genes. Gene structures are shown at the top of the figure, with exons represented as boxes and introns represented as cornered lines connecting the exons. Exons are shaded to represent the portion of the genes sequenced in the allelic survey. Line names are on the left and are grouped if they share the same haplotype of inactivating mutations among the four genes (i.e., single-nucleotide polymorphisms are not considered in these haplotype groupings). Downward-pointing triangles are insertions; upward-pointing triangles deletions; and associated numbers refer to the number of nucleotides in the indel. A premature stop codon is marked with an “X.” A complex indel involving the replacement of 37 bp with 19 different bases = an 18-bp net deletion and the creation of a premature stop codon is indicated with a star. A glycine-to-serine substitution in the oxyanion hole predicted to alter function is shown as a diamond. For some lines, we obtained two alleles (shown as double lines). The ratio of the average pairwise divergence of alleles at replacement (πa) and silent sites (πs) is shown below each gene
The frequency of null alleles in each of the four genes mirrors the selective constraint observed in the ratio of nonsynonymous-to-synonymous polymorphism in each gene. Specifically Est6b, which has no null alleles, exhibits a much lower ratio of nonsynonmous-to-synonymous polymorphisms per site (πn/πs = 0.08) than do the other three β-esterases (Est6a = 0.36, Est7 = 0.45, and Est6c = 0.50; see Supplementary Table 5). Combined, these data suggest that if these genes really are functionless, then they have become so since their respective alleles shared a common ancestor. Their inactivation could thus be very recent relative to the age of the divergence of the paralogous sequences.
To formally test whether the nonsynonymous-to-synonymous ratios differ for polymorphisms within a gene versus divergence between genes (paralogs or orthologs), we performed McDonald–Kreitman tests (McDonald and Kreitman 1991). If most of the polymorphisms observed had accumulated while the locus was a pseudogene, and most of the divergence occurred while the locus was an active gene, we may expect a significant McDonald–Kreitman test result (with πn/πs closer to one than Dn/Ds). Given that the D. ananassae Est6 genes are more similar to paralogs than orthologs, paralogs were used to obtain the divergence data for the McDonald–Kreitman test. We observed 18 nonsynonymous and 11 synonymous polymorphisms among Est6c alleles and 117 nonsynonymous site differences and 187 synonymous site differences between Est6c and Est6b. These values result yield a significant McDonald–Kreitman test result (G test p = 0.014), and the deviations occur in the direction expected if Est6c recently became a pseudogene (πn/πs = 0.5, Dn/Ds = 0.13).
Our interpretation is that most of the polymorphisms in Est6c have not had any selective constraint acting on them, whereas most of the polymorphisms in the other two genes (which had nonsignificant McDonald–Kreitman test results; synonymous polymorphisms/nonsynonymous polymorphism/synonymous divergence/nonsynonymous divergence Est6a:8/10/110/58 p = 0.12 and Est6b: 18/7/94/52 p = 0.50) have had selective constraint acting on them to an extent similar to that acting on the substitutions, which have become fixed since their divergence. To express this in another way, it is possible that Est6a and Est6b may have recently undergone a shift in selective pressure and that the polymorphism spectrum has not reached equilibrium. However, the signal for such a possibility is not showed by the McDonald–Kreitman test.
The frequency of null alleles at 3 of the 4 D. ananassae β-esterase loci is reminiscent of the data for Est7 locus from D. melanogaster. Balakirev and Ayala (2004) found 3 inactivating mutations (2premature termination codons and 1 frame-shifting indel) segregating in 17 of the 78 Est7 (they refer to this gene as Est6Ψ) sequences surveyed. The frequency of “inactivated alleles” varied with population: 11 of 28 North American alleles and 6 of 18 of the European sample were inactivated; however, none were found among the 12 African or 20 South American alleles. Despite the high frequency of null alleles, Balakirev and Ayala (2004) still found that nucleotide diversity at nonsynonymous sites (0.0076) was much lower than that at synonymous sites (0.0244), suggesting some kind of purifying constraint acting on this locus. This is similar to what we found in D. ananassae. Furthermore Dumancic et al. (1997) showed that some alleles of Est7 encode a protein with readily detectable esterase activity. We favour the interpretation that Est7 had a function that is no longer under strong selection in some populations of D. melanogaster. Similarly, the high frequency of null alleles in three of the four β-esterase loci of D. ananassae (including the ortholog of Est7 in D. melanogaster) suggests that the functional alleles provide only a marginal or perhaps temporally sporadic fitness advantage over the inactive alleles. Another interpretation suggested by Balakirev and Ayala (2004) is that Est7 might maintain a function that depends on a nonenzymatic, possibly regulatory, interaction with the neighbouring Est6.
Why So Many β-Esterase Gene Duplications and Losses?
The sequencing of the genomes of 12 Drosophila species has yielded important insights into the processes of gene gain and loss through genome-wide analysis (Hahn et al. 2007; Heger and Ponting 2007) and through studies of particular genes and gene families (Low et al. 2007; McBride et al. 2007). This study on the β-esterase loci details several important features of the molecular evolution that may apply generally to the processes of gene duplication and loss. Through reconciliation of the β-esterase gene number with the species tree, a minimum of 9 gain–loss events have occurred in this lineage since the ancestral species existed approximately 60 million years ago. Sequence-based phylogenies suggest more gene duplication events (11 to 12, depending on the method); however, these estimates must be qualified by hints of interparalog exchange, which may lead to overestimation of the number of gene duplication events. Even if we are conservative with our estimate of gene duplication in this cluster (i.e., 7 duplications), when we assume that the evolutionary time represented in the radiation of the 12 Drosophila species is approximately 400 million years (estimated by summing the time represented in all of the branches from Fig. 1 in Hahn et al 2007) and that the ancestral species had two β-esterase genes, then duplications of these genes have occurred much more often than estimated for the average gene (β-esterases: 7 duplications/2 ancestral genes/400 my = 0.009 duplications/gene/million years versus the 0.0012 duplications/gene/million years estimated to be the average by Hahn et al. 2007). Thus, Hahn’s metaphorical “revolving door” of gene gain–loss appears to be spinning faster in the β-esterase genes.
We are left with the question of why there might be so many gene gains and losses in the β-esterase gene cluster. It would seem that the requirements for the functions of these genes have fluctuated over evolutionary time, possibly in response to changes in environmental niches. However, the reproductive functions of some β-esterases suggest another possibility: the copy number changes are driven by sexual competition or conflict. Esterase 6 of D. melanogaster is transferred from the male ejaculatory duct to female flies on mating, and then it passes into the hemolymph and provokes female flies to lay rather than remate. In D. virilis, two β-esterase genes—the noncatalytic EstS gene and a catalytically active esterase (it is not clear which gene of the β-esterase cluster encodes it)—are expressed in the ejaculatory bulb and the catalytically active esterase, at least, is transferred to female flies on mating, although it does not move into the haemolymph. The high rate of duplications of the β-esterases and their reproductive functions concurs with the claims of Hahn et al. (2007) that the four gene families that had expanded substantially in the D. melanogaster lineage (the sperm-specific dynein [Sdic] family, two different serine-type peptidases, and protein kinases in the Stellate group associated with meiotic drive) may have a role in reproduction. Although expression in male reproductive tissue is not observed for D. yakuba and D. erecta Est6 or for D. pseudoobscura Est5b, it will be interesting to know whether any of the other hitherto uncharacterized β-esterases (such as the D. ananassae esterases) are expressed in these tissues and whether they affect remating and egg-laying behaviors.
References
Balakirev ES, Anisimova M, Ayala FJ (2006) Positive and negative selection in the beta-esterase gene cluster of the Drosophila melanogaster subgroup. J Mol Evol 62:496–510
Balakirev ES, Ayala FJ (1996) Is esterase-P encoded by a cryptic pseudogene in Drosophila melanogaster? Genetics 144:1511–1518
Balakirev ES, Ayala FJ (2003) Molecular population genetics of the beta-esterase gene cluster of Drosophila melanogaster. J Genet 82:115–131
Balakirev ES, Ayala FJ (2004) The beta-esterase gene cluster of drosophila melanogaster: Is psiEst6 a pseudogene, a functional gene, or both? Genetica 121:165–179
Balakirev ES, Chechetkin VR, Lobzin VV, Ayala FJ (2005) Entropy and GC content in the beta-esterase gene cluster of the Drosophila melanogaster subgroup. Mol Biol Evol 22:2063–2072
Brady JP, Richmond RC (1990) Molecular analysis of evolutionary changes in the expression of Drosophila esterases. Proc Natl Acad Sci U S A 87:8217–8221
Brady JP, Richmond RC (1992) An evolutionary model for the duplication and divergence of esterase genes in Drosophila. J Mol Evol 34:506–521
Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681
Campbell PM, Harcourt RL, Crone EJ, Claudianos C, Hammock BD, Russell RJ, Oakeshott JG (2001) Identification of a juvenile hormone esterase gene by matching its peptide mass fingerprint with a sequence from the Drosophila genome project. Insect Biochem Mol Biol 31:513–520
Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN et al (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218
Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR, Feyereisen R, Oakeshott JG (2006) A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol 15:615–636
Collet C, Nielsen KM, Russell RJ, Karl M, Oakeshott JG, Richmond RC (1990) Molecular analysis of duplicated esterase genes in Drosophila melanogaster. Mol Biol Evol 7:9–28
Dumancic MM, Oakeshott JG, Russell RJ, Healy MJ (1997) Characterization of the EstP protein in Drosophila melanogaster and its conservation in drosophilids. Biochem Genet 35:251–271
East PD, Graham A, Whitington G (1990) Molecular isolation and preliminary characterization of a duplicated esterase locus in Drosophila buzzatii. In: Barker JSF, Starmer WT, McIntyre RJ (eds) Ecological and evolutionary genetics in Drosophila. Plenum, New York, NY, pp 389–406
Enikolopov GN, Malevanchuk OA, Peunova NI, Sergeev PV, Georgiev GP (1989) The est locus of Drosophila virilis contains two related genes. Dokl Akad Nauk SSSR 306:1247–1249
Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413
Felsenstein J (1989) Phylip-phylogeny inference package. Cladistics 5:164–166
Gilbert MM, Auld VJ (2005) Evolution of clams (cholinesterase-like adhesion molecules): Structure and function during development. Front Biosci 10:2177–2192
Grisaru D, Sternfeld M, Eldor A, Glick D, Soreq H (1999) Structural roles of acetylcholinesterase variants in biology and pathology. Eur J Biochem 264:672–686
Hahn MW, Han MV, Han SG (2007) Gene family evolution across 12 Drosophila genomes. PLoS Genet 3:e197
Heger A, Ponting CP (2007) Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res 17:1837–1849
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Karotam J, Delves AC, Oakeshott JG (1993) Conservation and change in structural and 5’ flanking sequences of esterase 6 in sibling Drosophila species. Genetica 88:11–28
King LM (1998) The role of gene conversion in determining sequence variation and divergence in the Est5 gene family in Drosophila pseudoobscura. Genetics 148:305–315
Lachaise D, Cariou M-L, David JR, Lemeunier F, Ashburner M (1988) Historical biogeography of the D. melanogaster species subgroup. Evol Biol 15:9–226
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Low WY, Ng HL, Morton CJ, Parker MW, Batterham P, Robin C (2007) Molecular evolution of glutathione S-transferases in the genus Drosophila. Genetics 177:1363–1375
Martin DP, Williamson C, Posada D (2005) RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262
McBride CS, Arguello JR, O’Meara BC (2007) Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177:1395–1416
McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654
Meikle DB, Sheehan KB, Phyllis DM, Richmond RC (1990) localization and longevity of seminal-fluid esterase-6 in mated female Drosophila melanogaster. J Insect Physiol 36:93–101
Oakeshott JG, Boyce TM, Russell RJ, Healy MJ (1995) Molecular insights into an enzyme- esterase 6 in Drosophila. Trends Ecol Evol 10:103
Oakeshott JG, Claudianos C, Russell RJ, Robin GC (1999) Carboxyl/cholinesterases: A case study of the evolution of a successful multigene family. Bioessays 21:1031–1042
Oakeshott JG, Healy MJ, Game AY (1990) Regulatory evolution of the beta carboxyl esterases in Drosophila. In: Barker JSF, Starmer GA, McIntyre RJ (eds) Ecological and evolutionary genetics in Drosophila. Plenum, New York, NY, pp 359–387
Oakeshott JG, van Papenrecht EA, Boyce TM, Healy MJ, Russell RJ (1993) Evolutionary genetics of Drosophila esterases. Genetica 90:239–268
Oakeshott JG, van Papenrecht EA, Claudianos C, Morrish BC, Coppin C, Odgers WA (2000) An episode of accelerated amino acid change in Drosophila esterase-6 associated with a change in physiological function. Genetica 110:231–244
Olson PF, Fessler LI, Nelson RE, Sterne RE, Campbell AG, Fessler JH (1990) Glutactin, a novel Drosophila basement membrane-related glycoprotein with sequence similarity to serine esterases. EMBO J 9:1219–1227
Pearson WR (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol 183:63–98
Pen J, Van Beeumen J, Beintema JJ (1986) Structural comparison of two esterases from Drosophila mojavensis isolated by immunoaffinity chromatography. Biochem J 238:691–699
Posada D, Crandall KA (1998) MODELTEST: Testing the model of DNA substitution. Bioinformatics 14:817–818
Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R (2002) Evolution of supergene families associated with insecticide resistance. Science 298:179–181
Richmond RC, Nielsen KM, Brady JP, Snella EM (1990) Physiology, biochemistry and molecular biology of the Est6 locus in Drosophila melanogaster. In: Barker JSF, Starmer WT, McIntyre RJ (eds) Ecological and evolutionary genetics of Drosophila. Plenum, New York, NY, pp 273–293
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: Sequence visualization and annotation. Bioinformatics 16:944–945
Swofford DL (2003) PAUP*. Phylogenetics analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA
Saad M, Game AY, Healy MJ, Oakeshott JG (1994) Associations of esterase 6 allozyme and activity variation with reproductive fitness in Drosophila melanogaster. Genetica 94:43–56
Sergeev PV, Panin VM, Pavlova GV, Kopantseva MR, Shostak NG, Bashkirov VN, Georgiev GP, Korochkin LI (1995) The expression of esterase S gene of Drosophila virilis in Drosophila melanogaster. FEBS Lett 360:194–196
Sergeev PV, Yenikolopov GN, Peunova NI, Kuzin BA, Khechumian RA, Korochkin LI, Georgiev GP (1993) Regulation of tissue-specific expression of the esterase S gene in Drosophila virilis. Nucleic Acids Res 21:3545–3551
Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN et al (2007) Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450:219–232
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamarina NA, Ludwig MZ, Richmond RC (1997) Divergent and conserved features in the spatial expression of the Drosophila pseudoobscura esterase-5B gene and the esterase-6 gene of Drosophila melanogaster. Proc Natl Acad Sci U S A 94:7735–7741
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917
Acknowledgements
This work was supported in part by a grant from the Australian Research Council to CR (DP0557497). We thank an anonymous reviewer for constructive criticism.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Robin, C., Bardsley, L.M.J., Coppin, C. et al. Birth and Death of Genes and Functions in the β-Esterase Cluster of Drosophila . J Mol Evol 69, 10–21 (2009). https://doi.org/10.1007/s00239-009-9236-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9236-3