Abstract
Coral propagation and out-planting are becoming commonly adopted as part of reef stewardship strategies aimed at improving reef resilience through enhanced natural recovery and rehabilitation. The coral microbiome has a crucial role in the success of the coral holobiont and can be impacted shortly after out-planting. However, long-term characterisation of the out-plant microbiome in relation to out-plant survival, and how these properties vary across reef sites, is unexplored. Therefore, at three reef sites on Opal Reef, Great Barrier Reef (Mojo, Sandbox and Rayban, 16°12′18″S 145°53′54″E), we examined bacterial communities associated with out-planted Acropora millepora coral and monitored coral survival over 12 months (February 2021–22). Bacterial communities of out-planted corals exhibited significant changes from donor colonies 7 days to 1.5 months after out-planting. Further, bacterial community composition differed for sites Sandbox and Rayban with low overall survival (0–43%) versus Mojo with higher overall survival (47–75%). After initial dissimilarity in bacterial communities of out-plants across sites at 1.5 months, and despite changes within sites over time, out-plants exhibited similar microbial communities across sites at 7 days and 6, 9 and 12 months. We hypothesise these trends reflect how bacterial communities are shaped by rapid changes in local environmental characteristics (e.g. from source to out-planting site), where out-plant bacterial communities ‘conform’ to out-planting site conditions. After initial changes, out-plant bacterial communities may then be under the influence of global environmental conditions—such as annual trends in temperature across seasons. Such outcomes indicate the importance of site selection in shaping initial coral bacterial communities and subsequent out-plant success. Importantly, continued differences in out-plant survival trajectory but similar bacterial communities across sites after 1.5 months indicate that other factors—apart from bacterial community changes—likely govern out-plant success in the longer term. Our research highlights the need to resolve drivers of small-scale site differences alongside higher resolution spatiotemporal monitoring of environmental conditions to distinguish key drivers of (i) microbial change during out-planting and (ii) out-plant survival to subsequently inform out-plant site selection to optimise future restoration efforts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Increasingly frequent and intense anthropogenic disturbances threaten the persistence of tropical coral reefs and hence the wealth of ecological, economic, social and heritage values they provide (Hughes et al. 2017; Eakin et al. 2019; Eddy et al. 2021; Sully et al. 2022). Future survival of coral reef ecosystems is primarily reliant on mitigation of climate change to reduce global stress from ocean warming, acidification and deoxygenation (Kleypas et al. 2021; Shaver et al. 2022). However, parallel application of strategic, active management approaches will be pivotal for sustaining resilience of coral reef ecosystems and safeguarding against future climate conditions (Duarte et al. 2020; Kleypas et al. 2021; Shaver et al. 2022). Increasing application of active management approaches has accelerated over the last two decades in almost all tropical coral-reef-associated regions, whereby coral propagation and out-planting techniques have been utilised to rapidly increase coral biomass and cover at degraded or high-value reef sites (Boström-Einarsson et al. 2020; Hein et al. 2021), including on the Great Barrier Reef (Howlett et al. 2022; McLeod et al. 2022).
The ultimate success of coral propagation and out-planting rests on the capacity to rapidly up-scale and optimise approaches (Boström-Einarsson et al. 2020; McAfee et al. 2021; Suggett and van Oppen 2022). Whilst the readiness of successful implementation of different active management approaches is variable (Suggett and van Oppen 2022), in-water coral propagation and out-planting have become one of the most promising methods for local and targeted recovery or rehabilitation (Williams et al. 2019; Hein et al. 2020; Suggett et al. 2019). Even so, the long-term survival of corals during the propagation and out-planting process is highly variable, ranging from < 5% to > 90% (Boström-Einarsson et al. 2020; Ware et al. 2020; Suggett et al. 2019, 2020). Many factors can contribute to success in coral out-planting, such as out-planting technique (method of attachment), coral species, fragment size, reef area (back-reef, fore-reef vs reef crest) and geographical subregion (Bayraktarov et al. 2016; van Woesik et al. 2021; Boström-Einarsson et al. 2018; Suggett et al. 2019), but these are often site- and context-specific. Consequently, investigating how environmental factors and biological factors of the holobiont interact to influence out-plant survival is critical to long-term success of active management approaches that are increasingly necessary to ensure the long-term viability of reef ecosystems (Hein et al. 2020).
Coral microbiomes are an essential component of the coral holobiont (Bourne et al. 2016; Peixoto et al. 2017; Voolstra et al. 2021). However, only recently has the coral microbiome been considered as an important element in the success of active management approaches (van Oppen and Blackall 2019; Moriarty et al. 2020; Peixoto et al. 2021; Voolstra et al. 2021; Strudwick et al. 2022). For example, coral species can display community restructuring or stability during long-term (6 months) nursery propagation and subsequent early out-planting (1 day–1 month) (Strudwick et al. 2022). However, the longer-term dynamics of the bacterial communities associated with out-planted coral after relocation to the reef environment—and hence the role of bacterial communities influencing the success of newly out-planted coral—remains unexplored. Addressing such gaps may be critical where coral diseases have potential to impact survival during propagation and out-planting efforts (Moriarty et al. 2020). In fact, how microbes impact the survival of coral may be particularly important at degraded reef sites that are often targeted for out-planting, where there is potential for copiotrophic microorganisms, many of which are known coral pathogens, to be present in higher abundances (Dinsdale et al. 2008; Kelly et al. 2014; Haas et al. 2016; Silveira et al. 2017).
Opal Reef, on the northern Great Barrier Reef (GBR), has been a site of intensive propagation and out-planting activity since the 2018 initiation of the Coral Nurture Program, a research-led reef stewardship approach (Howlett et al. 2022). Relatively high survivorship (typically > 80%) has been recorded for Acropora spp.; however, survivorship remains highly variable across sites ranging from 79.8 to 100% (Suggett et al. 2020; Howlett et al. 2022). Considering that environmental conditions are known to drive bacterial community shifts (Kelly et al. 2014; McDevitt-Irwin et al. 2017; Maher et al. 2019; Camp et al. 2020), we hypothesised that (i) out-planted fragments will undergo changes in the bacterial community composition after out-planting, and that (ii) these changes will differ between reef sites. Furthermore, given that associated bacterial communities are essential for coral host health (Bourne et al. 2016; Peixoto et al. 2017; Voolstra et al. 2021), we hypothesise that (iii) shifts in coral-associated bacterial communities are likely to reflect differences in survivorship. To test these hypotheses and inform future out-planting efforts, over 12 months, we examined temporal dynamics of coral-associated bacterial communities and survivorship of Acropora millepora fragments out-planted across three sites on Opal Reef (northern GBR) characterised by different defining features and environmental conditions.
Materials and methods
Sampling location and experimental design
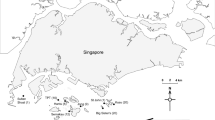
To examine differences in coral-associated bacterial communities amongst coral fragments out-planted at each reef site with contrasting characteristics, we performed a transplantation experiment using the coral species A. millepora. This species is routinely grown in coral nurseries and used for out-planting on the GBR (e.g. Howlett et al. 2022) and exhibits microbiome variability over time and space during propagation in coral nurseries at Opal Reef (Strudwick et al. 2022). Experiments were conducted at three sites across Opal Reef (16°12′18"S 145°53′54"E), which is a 24.7 km2 reef situated on the northern GBR (detailed in Suggett et al. 2019; Howlett et al. 2021) (Fig. 1.). Each site had contrasting characteristics: (i) “Mojo”, subject to strong tidal currents due to its close proximity to a deep-water channel leading to the coral sea at the north of Opal Reef; (ii) “Rayban”, is not subject to strong currents due to its central location at Opal Reef within a sheltered sandy lagoon area (see Suggett et al. 2019; Howlett et al. 2021), and (iii) “Sand Box”, is adjacent to a channel on the southern edge of Opal reef and consequently has elements of both sites with some sheltered sandy lagoons and mild currents (Edmondson personal obs.).
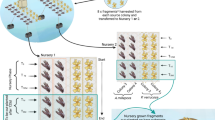
In February 2021, coral fragments (~ 5–10 cm) were harvested from five established A. millepora nursery colonies at Rayban for out-planting across five separate “plots” within each of the three sites (Fig. 2a. and Fig. S2.). Five nursery colonies (~ 40 cm diameter) were selected from two adjacent established (2 years) nursery frames. From each donor colony, ~ 60 fragments (standardised to sizes between 5 and 10 cm) were harvested. Two fragments (≤ 5 cm) were randomly selected after the donor colony was fragmented and were retained to capture any potential heterogeneity in bacterial communities across the colony (Marchioro et al. 2020; Damjanovic et al. 2020)—in a sterile zip-lock bag, returned to the operations vessel and preserved in RNAlater for microbiome characterisation (defined as “TF”—time of fragmentation). The other 58 fragments were held in wire trays on the nurseries at Rayban for 24–48 h—to enable identification of any fragments that exhibited mortality from fragmentation and also because of sporadic access to the three sites prior to out-planting; specifically, corals out-planted at Rayban were held for 24 h, whilst those out-planted at Sandbox and Mojo were held for 48 h prior to out-planting.
a Experimental set-up: illustrating the process of fragmentation (TF) for one donor colony from the nursery through to out-planting (TO-1 at 24 h and TO-2 at 48 h) at one plot within the three reef sites. This was repeated for the remaining four donor colonies. b Sampling: illustrating the process of one sampling time point, at each reef site the five out-plant plots were sampled five times*over a 12-month period. c Timeline: illustrating the points at which sampling occurred, at TF/O only the donor colonies from the nursery frames were sampled and at subsequent sampling points only out-plants were sampled T7d-12 m. *Unless there was total mortality at the plot prior to completion of the experiment
At each reef site, 11–23 fragments from each donor were out-planted within five separate plots (Fig. 2a, b. and Table S1.). Out-planting plots were approximately 4–5 m apart and within an area of ≥ 2 m2 (Fig. S2) and marked with cattle tags to identify the donor colony they were planted from. Each plot satisfied four pre-requisites: (i) bare consolidated structurally sound substrate (i.e. not rubble or sand), (ii) absence of algal turfs, (iii) located outside of damselfish territories, (iv) presence of other coral growing within a 1–2 m radius. Coralclip® (a stainless-steel spring-clip that is fastened using a hammer and masonry nail) was used to fasten each fragment to the substrate (as per Suggett et al. 2020). At the time of initial out-planting (24 h: TO-1 and 48 h: TO-2), two fragments (≤ 5 cm) from each donor colony were retained and preserved in the same manner as for TF (above) for microbiome characterisation (Fig. 2a.). This was to ensure adequate representation of the fragments at time of out-planting, and to assess whether the bacterial communities changed from time of fragmenting (TF) to time of out-planting (TO-1 and TO-2). Once all fragments were out-planted, photographs were taken of the entire plot and the out-plants were counted. Fragment survivorship and coral-associated bacterial communities of out-planted fragments were then tracked over 12 months.
Coral sample preservation and DNA extraction
Following TF, TO-1 and TO-2, one out-plant was sampled from each plot and site (5 plots × 3 sites = 15 fragments total) at seven days (T7d), 1.5 months (T1.5 m), six months (T6m), nine months (T9m) and 12 months (T12m) (Fig. 4b, c). At each time point, fragments (≤ 5 cm) were subsampled from coral out-plants using wire clippers. After sampling, all fragments were returned to the operations vessel (Wavelength IV) in sterile zip-lock bags within 30–50 min, placed into sterile 15 mL falcon tubes and preserved by total submersion in RNAlater. All samples were subsequently held at ambient temperature for 6 days during transportation from the study site to the laboratory for processing (as per Strudwick et al. 2022). One donor colony sample was compromised during transit and consequently one replicate for donor colony five at TF proceeded to subsequent DNA extraction and sequencing. Once in the laboratory, RNAlater was thoroughly removed from 15 mL falcon tubes using an adjustable pipette with sterile tips, after which samples were preserved at – 80 ℃ for 1–11 months and DNA extractions were all conducted at the same time. Prior to DNA extraction, coral tissue was removed from the coral skeleton, using an air brushing technique. Coral fragments were thawed on ice in their respective 15 mL falcon tubes, removed from the falcon tube using sterile forceps, rinsed with autoclaved phosphate-buffered saline (PBS) (3X, pH 7.4) to remove RNAlater residue, placed in sterile zip-lock bags and air brushed with sterile pipette tips into 4 mL of autoclaved PBS (3X, pH 7.4). The tissue slurry was divided across two 2 mL micro centrifuge tubes and centrifuged at 8000 rpm for 5 min. The supernatant was removed, and the pellet was stored at – 80 °C for 4–6 weeks until DNA extraction.
DNA was extracted from approximately 100 µL of the pellet using a DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s protocol (July 2020 version) with a total elution volume of 40 µL. Extractions were conducted in randomised batches of 23 with one kit negative sample included. Extracted DNA was quality checked and the concentration was quantified using a NanoDrop spectrophotometer.
Environmental logging
Upon initiation of the experiment, temperature, light attenuation and flow at each reef site were measured using HOBO™ loggers for a 14-day period to identify potential site-specific differences in key environmental factors that can influence coral growth and survivorship. Due to site access, sensors could only be deployed for a 14-day period. Whilst this is a relatively short timeframe, it allowed a spring-neap cycle to be captured to identify relative differences across the sites. Light attenuation was measured by securing two temperature/light data loggers (HOBO Pendant® UA-002–64), positioned 1 m apart onto a PVC pipe with metal brackets and vertically suspended the structure in the water column. Each logger was secured in a horizontal orientation so that the light sensors were parallel to the surface and did not shade one another (Fig. S1a-b.). Light sensors were routinely cleaned (approximately every three days) to prevent biofouling. Light and temperature data were recorded hourly and temperature readings were taken from only the bottom data logger on the PVC pipe set-up. All temperature/light data loggers were inter-calibrated with data from a 4 h period of logging at 10-min intervals in the same position and conditions. Average temperature and light intensity of the three loggers were calculated at each 10-min interval. Relative error was calculated for individual loggers at every 10-min interval during the calibration period. Light and temperature readings recorded at each site during the 14-day monitoring period were then adjusted by their logger’s average relative error calculated during the calibration. Light attenuation was calculated using the following equation (Kirk 1994):
la is the PAR recorded by the deepest logger and lb is the PAR recorded by the shallower logger (separated by a 1 m interval), and k is the attenuation coefficient (m−1).
Flow was recorded using a G data logger (HOBO Pendant® UA-004–64) that was assembled into a tilting current metre (based on Crookshanks 2008, see Supplementary Appendix 1 for detailed description): The tilting current metres were cross-calibrated and flow readings ranged from low (0) to high (1), as calibration to a known flow rate (m/s−1) was not possible (Fig. S1f.). To inter-calibrate tilting current metres, for a 24 h period, all tilting current metres were secured to an in situ nursery frame within 30 cm of each other (Fig. S1c) with G data loggers recording at 20-min intervals. Average flow was calculated at each 20-min interval. Relative error was calculated for individual tilting current metres at every 20-min interval during the calibration period. Recordings from each site during the 14-day monitoring period were then adjusted by their logger’s average relative error calculated during the calibration (calibrated flow, temperature and light attenuation data provided as Supplementary Data S1).
Environmental sampling and DNA extraction
Water and substrate samples were collected to characterise environmental bacterial communities prior to introduction of coral material and were only collected at time of initial out-planting, as such it was not possible to characterise any potential variability in substrate and water communities over time. Water samples for microbiome characterisation were collected from the surface at each reef site using 10 L pre-sterilised plastic containers and filtered in triplicate through 47 mm, 0.22 µm pore-size membrane filters (Millipore, DURAPORE PVDF 0.22 µm WH PL) using a peristaltic pump (100 rpm), within 10 min of sample collection. Before each sample was filtered, 250 mL 10% bleach was run through the pump, followed by 500 mL MilliQ water, and then 3 L of sample. Filters were stored in cryovials, snap-frozen and stored in a dewar for transport back to the laboratory where they were stored at – 80 °C. DNA was extracted from the membrane filters using the PowerWater DNA isolation Kit (QIAGEN) as per the manufacturer’s protocol (June 2016 version). DNA extractions were performed alongside three kit blanks, which were subsequently included in all sequencing analysis to exclude kit contaminants.
Substrate samples for microbiome characterisation were also collected using a hammer and chisel to leverage a 2 cm × 2 cm × 2 mm piece of substrate off each plot into a sterile zip-lock bag. Following sampling, all substrate samples were preserved, stored and transported to the laboratory in the same manner as coral samples. Once in the laboratory, samples were preserved at 4 °C for 11 months until RNAlater was thoroughly removed from 15 mL falcon tubes using an adjustable pipette with sterile tips, after which DNA was extracted. DNA was extracted from approximately 100 µg of the substrate sample using a DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s protocol (July 2020 version) with a total elution volume of 40 µL. Extractions were performed alongside one kit negative sample and the kit negative sample was included in subsequent sequencing analysis to exclude kit contaminants. All extracted DNAs were quality-checked, and the concentration was quantified using a NanoDrop spectrophotometer.
Overall, 126 samples were collected for bacterial community analysis: (i) out-plants: three sites × three-five plots (some plots experienced total mortality) × five time points (n = 64), (ii) donor colonies: two fragments x five donor colonies × TF plus two fragments x five donor colonies × two out-planting times TO (n = 30), (iii) environmental: three sites × three replicate water samples (n = 9) and three sites × five plots × one substrate sample (n = 15) plus eight x DNA extraction blank samples.
Quantification of coral out-plant survival
Coral survival was recorded at each sampling time point (T7d, T1.5 m, T6m, T9m, T12m) and through additional opportunistic observations at 35, 40, 167, 344 and 346 days after out-planting. Survival was defined as the number of fragments still alive in the out-plant plot at each time interval as a percentage of the number out-planted. Corals in the out-plant plot observed with < 5% live tissue were counted as “dead”, whilst fragments either missing or dislodged from the Coralclip® were defined as “detached”. Corals sampled for preservation at time of survival counts were included in “live” counts. Both dead and detached coral fragments were considered as “lost” and excluded from survival counts.
16S rRNA amplicon sequencing
Extracted DNA was stored at – 80 ℃ for two weeks prior to 16S rRNA amplicon sequencing, which was used to characterise the composition and diversity of coral, sediment, water and kit negative bacterial communities. The hypervariable V3 and V4 regions of the bacterial 16S rRNA gene were amplified using the primers 341F (5’–CCTAYGGGRBG-CASCAG-3’) and 805R (5’–GACTACHVGGGTATC-TAATCC-3’) (Klindworth et al. 2013), prior to sequencing on the Illumina MiSeq platform (Ramaciotti Centre for Genomics (Sydney, NSW, Australia)). Raw data files in FASTQ format were deposited in NCBI Sequence Read Archive (SRA) under Bioproject number PRJNA929655.
Bioinformatics
Raw demultiplexed sequencing data were analysed with Quantitative Insights into Microbial Ecology (QIIME 2, version 2020.6) platform (Callahan et al. 2016). The DADA2 plugin was used to denoise the data (Callahan et al. 2016) and taxonomy was assigned using the classify-sklearn classifier (Pedregosa et al. 2011) against the SILVA v138 database. In total, 9,156,785 reads were generated (after denoising) from 126 samples. Amplicon sequence variants (ASVs) corresponding to chloroplast or mitochondria were filtered from the data set. 18 ASVs that comprised 51% of sequences in the DNA extraction negative controls and have been previously reported as contaminants of laboratory reagents (Weyrich et al. 2019), were removed for subsequent analyses using the filter command in R (version 4.2.2). Prior to diversity analyses, four samples were removed from the data set due to poor sequencing outputs leading to low read numbers after quality filtering and contaminant removal (< 900 reads or < 58 ASVs). As a result of filtering and complete mortality at some plots, six time points had three biological replicates and one time point had four biological replicates, all other time points had five biological replicates. For beta diversity analyses, the raw read ASV table was converted to relative abundances, scaled to 20,000 (McKnight et al. 2019) and square root-transformed.
Statistical analysis
Differences in bacterial community structure and dispersion (beta diversity patterns) of out-planted corals between reef sites at each time point (reef site = fixed effect), between donor colonies at time of fragmenting (TF) and out-planted corals over the 12-month time period within a site and over time—from 7 days to 1.5 months (T7d-T1.5 m), 1.5 months to 6 months (T1.5 m-T6m), 6 months to 9 months (T6m-T9m) and 9 months to 12 months (T9m-T12m)—were analysed using the Bray–Curtis dissimilarity distance metric. Differences in bacterial community structure of water and substrate between reef sites were also analysed using the Bray–Curtis dissimilarity metric. Permutation tests for homogeneity in multivariate dispersion (PERMDISP) of coral-associated bacterial community were calculated using the betadisper function of the ‘vegan’ R package. Patterns in bacterial community structure were visualised using non-metric multidimensional scaling (nMDS) plots. Differences in community structure were tested for significance with pairwise permutational multivariate analysis of variance (PERMANOVA; perm = 999) of Bray–Curtis dissimilarities using the pairwise.adonis function of vegan and differences in community dispersion were tested for significance—with site as a single factor and with pairwise comparisons between sites at each timepoint—using the permutest.betadisper function of vegan (perm = 999), p values were adjusted by applying a Benjamini and Hochberg (a.k.a. False Discovery Rate) correction, all padj values < 0.05 were considered significant. When significant differences in the bacterial community structure were identified, similarity percentage analysis (SIMPER) was used in PAST 4.03 to identify and calculate the percentage contribution of each ASV to dissimilarity between groups, ASVs contributing > 1.5% dissimilarity were reported. The core_members function of the ‘microbiome’ R package was used to identify core bacterial community members (present in > 80% samples with relative abundance > 0.1%) for out-planted fragments over time and at time of fragmenting (TF).
Kaplan–Meier survival curves were estimated for out-plant survival at each site. To test for differences in survival between sites, we conducted a pairwise log-rank test, p values were adjusted by applying Bonferroni adjustment. To assess differences between sites in flow, attenuation and temperature during the 14-day monitoring period, each variable was analysed separately and sites were compared via a Kruskal–Wallis tests to assess significance, when significant (p < 0.05), Dunn’s post hoc test was applied with subsequent Bonferroni adjustment, all padj values < 0.05 were considered significant. To qualitatively describe overall similarity of sites (from the recorded parameters), a dissimilarity matrix of the mean temperature, light attenuation and flow was used (Table S2.).
Results
Site-specific environmental characteristics and microbial communities
Flow rates, light attenuation and temperature differed between the three reef sites during the 14-day monitoring period (Kruskal–Wallis test, p < 0.001, Supplementary Data S2). All sites exhibited different variance for flow, light attenuation and temperature (Dunn’s post hoc, padj < 0.001, Supplementary Data S2). The largest range in light attenuation was at Mojo (1.54 m−1), followed by Rayban and Sandbox (1.52 m−1 and 1.11 m−1 respectively, Fig. S3b.), and the highest mean light attenuation was at Mojo, followed by Rayban and Sandbox (0.415, 0.317 and 0.200 m−1 respectively, Fig. S3b.). The largest range in flow was also observed at Mojo (1.20), followed by Sandbox and Rayban (1.13 and 0.58 respectively, Fig. S3a.), and similarly, the highest mean flow was observed for Mojo, followed by Sandbox and Rayban (0.455, 0.335 and 0.240 respectively, Fig. S3a.). The largest range in temperature was observed at Rayban (2.91 ℃) followed by Mojo and Sandbox (2.41 ℃ and 1.91 ℃ respectively, Fig. S3c.). The highest mean temperature was observed at Sandbox (31.61 ± 0.42 ℃) followed by Rayban (30.36 ± 0.54 ℃) and Mojo (29.80 ± 0.44 ℃) (Fig. S3c.). In summary, Mojo was characterised by the highest mean flow, the lowest water clarity (the highest mean light attenuation), the lowest mean temperature, and the largest range in temperature and flow. Sandbox had intermediate mean flow and the highest temperature, but the highest water clarity (lowest light attenuation) and the smallest range in light attenuation and temperature. Rayban had intermediate mean temperature, the lowest mean flow, intermediate water clarity and the highest range in temperature. Overall, Rayban and Mojo were more similar than Rayban and Sandbox based on a qualitative comparison of the recorded parameters (Table S2).
Bacterial community composition differed between water and substrate within every site (PERMANOVABray-Curtis padj < 0.05, Figs. S4–5 and Supplementary Data Sheet S3.), reflecting predominantly different environmental bacterial communities. However, water samples were generally the same between sites (PERMANOVABray-Curtis, padj > 0.05, Supplementary Data S3.), whereas substrate samples associated with Sandbox differed with those from Rayban (PERMANOVABray-Curtis, F = 1.403, padj = 0.021, Figs. S4–5., and Supplementary Data S3). These differences in substrate bacterial communities between Rayban and Sandbox occurred in parallel with differences between the two sites in measured mean flow, temperature and light attenuation. However, there were no further differences in substrate bacterial communities between sites despite flow, temperature and light attenuation also differing between Rayban and Mojo, and Sandbox and Mojo.
Coral-associated bacterial communities changed after out-planting across three reef sites
Bacterial community composition associated with the donor colonies at time of fragmenting (TF) was first compared to that at initial out-planting (TO-1 and TO-2) to account for any possible responses induced by the different periods of time with which fragmented material was held prior to out-planting (TO-1: 24 h Rayban vs TO-2: 48 h Sandbox, Mojo). No changes were observed from TF vs TO for any site (both TO-1 and TO-2) (PERMANOVABray-Curtis, padj < 0.05, Supplementary Data S6.), and hence subsequent comparisons between donor colonies and out-planted fragments were conducted against samples from time of fragmenting (TF) to ensure consistency between sites. After out-planting, bacterial communities associated with the corals at all sites became significantly different to those at time of fragmenting (TF), this occurred 1.5 months after out-planting at Mojo and Rayban (PERMANOVABray-Curtis, F = 2.250, padj = 0.009, df = 1; F = 2.571, padj = 0.002, df = 1 respectively), but after only 7 days at Sandbox (PERMANOVABray-Curtis, F = 2.501, padj = 0.017, df = 1, Fig. 3a–c, Fig. 4 and Fig. S6a–c). As such, shifts in bacterial community composition were more rapid at Sandbox compared to both Rayban and Mojo.
Bacterial community structure and relative dispersion of the microbial communities at time of fragmenting (TF) and out-planted fragments at a Rayban, b Sandbox and c Mojo over time. Plots are based on non-metric multidimensional scaling (n-MDS) of Bray–Curtis distances of bacterial community structure.
Bacterial community composition (relative abundances) by genus at time of fragmenting (TF) and of out-planted fragments over the 12-month monitoring period across the three sites. Pastel colours represent genera with an average relative abundance of < 0.1% in all samples, full legend provided as supplemental data (Supplementary Data S4)
One ‘core’ ASV was present at time of fragmenting (TF) and classified as a member of the Endozoicomonas genus (mean RA = 50.3%). Following significant changes in the bacterial community of out-plants at 7 days and 1.5 months after out-planting (Sandbox and Mojo/Rayban respectively), the core ASV from the Endozoicomonas genus (identified at TF) was only retained as a core ASV at Sandbox and Rayban and was completely lost from three of five replicates at Mojo and no longer classified as a core member in out-plants. In fact, whilst still classified as a core ASV, the Endozoicomonas ASV declined in relative abundance from time of fragmenting (TF mean RA = 50.3%) after 7 days at Sandbox (T7d mean RA = 22.6%) and 1.5 months at Rayban (T1.5 m mean RA = 8.4%) (Fig. S7).
Temporal dynamics of out-plant-associated bacterial communities varied across sites
At Mojo, there were significant changes in the bacterial community structure from 7 days (T7d) to 1.5 months (T1.5 m) post-out-planting after which the bacterial communities remained stable until a change in the out-plant bacterial community structure from 9 to 12 months (T9m-12 m) (PERMANOVABray-Curtis, padj = 0.031, F = 1.497, df = 1, and padj = 0.037, F = 1.432, df = 1 respectively). Differences in bacterial communities of out-planted fragments at Mojo from 7 days to 1.5 months (T7d-T1.5 m) were primarily explained by (> 1.5% dissimilarity contribution) decreased relative abundance (RA) the (TF) core Endozoicomonas ASV, increased abundance of 5 ASVs from the Lactobacillus fermentum, Ruegeria, Pseudovibrio and Erthryobacter genera and loss of an ASV from the Tenacibaculum genus (SIMPER, Supplementary Data S6.). Differences in bacterial communities of out-planted fragments at Mojo from 9 to 12 months (T9m-T12m) were explained by increased RA of the (TF) core Endozoicomonas ASV (Fig. S7) and four ASVs from the Endozoicomonas, Ruegeria, Psychrobacter and Synechococcus_CC9902 genera and increased RA of an ASV from the Hungateiclostridiaceae family (SIMPER, Supplementary Data S6). Bacterial communities of out-planted fragments also exhibited restructuring over time at Rayban. At Rayban, the coral-associated bacterial community structure of out-plants changed from 7 days (T7d) to 1.5 months (T1.5 m), and from 1.5 months (T1.5 m) to 6 months (T6m) after out-planting (PERMANOVABray-Curtis, padj = 0.032, F = 1.610, df = 1, and padj = 0.048, F = 1.247, df = 1 respectively). There were no further changes in coral out-plant bacterial community structure after 6 months (PERMANOVABray-Curtis, padj > 0.05). Similar to Mojo, differences in bacterial communities of out-planted fragments at Rayban from 7 days to 1.5 months (T7d-T1.5 m) were also primarily explained by decreased relative abundance (RA) of the (TF) core Endozoicomonas ASV and one other ASV from the Endozoicomonas genus (SIMPER, Supplementary Data S6.). Differences in bacterial communities of out-planted fragments from 1.5 months to 6 months (T1.5 m-T6m) at Rayban were explained by further decreases in RA of the two Endozoicomonas ASVs (Fig. S7) and increases in RA of ASVs from the Limnothrix, and Thrichodesmium_IMS101 genera (SIMPER, Supplementary Data S6.). After the initial change in bacterial community structure from time of fragmenting (TF) to 7 days after out-planting (T7d) at Sandbox, there were no further changes in the bacterial communities of out-planted coral fragments (PERMANOVABray-Curtis, padj > 0.05, Fig. S6. and Supplementary Data S6.). There were no changes in the dispersion of coral-associated bacterial communities for out-plants over the 12 months at any site (PERMUTEST, padj > 0.05, Fig. S8 and Supplementary Data S7).
Out-plant survival and associated bacterial communities varied across sites
Mean survival of out-plants at the final time point of sampling (12 months after out-planting) was higher at Mojo (mean 58.96% ± 1.72, n = 5) than Rayban and Sandbox (31.75% ± 6.25, n = 4 and 32.66% ± 7.31, n = 3). Throughout the experiment, mean survival remained 3.85–32.59% higher at Mojo compared to both other sites. Kaplan–Meier survival curves were significantly different between Mojo and Rayban (Pairwise log-rank, padj < 0.001) and Mojo and Sandbox (Pairwise log-rank test, padj < 0.0001, Fig. 5a.).

Source data are provided as Supplementary Data S5. †Indicates a significant difference in microbial community structure between Mojo and Rayban, and Mojo and Sandbox (PERMANOVABray-Curtis, padj < 0.05)
a Kaplan–Meier survival curve for out-plants at Mojo, Sandbox and Rayban, b bacterial community structure and relative dispersion of the microbial communities of out-planted fragments at Mojo, Sandbox and Rayban at individual time points and c photographs showing growth of a singular tracked fragment over time. Plots are based on non-metric multidimensional scaling (n-MDS) of Bray–Curtis distances of bacterial community structure.
Bacterial community composition of out-planted corals only significantly differed for Mojo vs Rayban and Mojo vs Sandbox at 1.5 months (T1.5 m) (PERMANOVABray-Curtis, F = 1.346, padj = 0.048, df = 1; F = 1.232, padj = 0.048, df = 1 respectively) at all other time points, and there were no differences across sites in the structure of the coral-associated bacterial communities (PERMANOVABray-Curtis padj > 0.05, Fig. 5b and Supplementary Data S6). Differences in bacterial communities of out-planted fragments between sites were primarily explained by higher RA at Sandbox and Rayban compared to Mojo of two ASVs from the genus Endozoicomonas (one of which was from the ‘core’ microbiome characterised at TF), as well as a lower relative abundance at Sandbox and Rayban of ASVs from the Lactobacillus, Ruegeria, Erythrobacter and Pseudovibrio genera compared to Mojo (SIMPER, and Supplementary Data. S6). The out-plant-associated bacterial communities were more heterogenous at Mojo compared to out-plants at Rayban at 1.5 months and 6 months (PERMUTEST, padj < 0.05, Fig. S8. and Supplementary Data S7) and compared to out-plants at Sandbox at 6 months and 9 months (PERMUTEST, padj < 0.05, Fig. S8. and Supplementary Data S7.).
Discussion
Active reef management approaches that include the translocation of corals within reef systems to enhance natural recovery are expanding on the Great Barrier Reef (McLeod et al. 2022; Howlett et al. 2022) and worldwide (Boström-Einarsson et al. 2020). However, some evidence indicates that coral-associated bacterial communities are impacted by transplantation (Casey et al. 2015; Ziegler et al. 2019; Haydon et al. 2021), as well as nursery-based propagation and out-planting (Strudwick et al. 2022), and it has been proposed that these changes are driven by exposure to novel environmental conditions between propagation sites (Strudwick et al. 2022). Even so, to date, there has been little quantification and comparison of long-term out-plant survival and associated bacterial community composition across reef sites with contrasting environmental conditions on the GBR. Most studies have only quantified survival of out-planted Acroporid spp. (van Woesik et al. 2018, 2021; Howlett et al. 2022) or monitored bacterial communities of out-planted corals for short periods of time (1–30 days, Strudwick et al. 2022). Here we show that on the northern GBR reef environmental conditions, out-plant survival, and coral-associated bacterial communities all exhibit inter-site variability. Specifically, A. millepora fragments from known donor colonies out-planted across three diverse reef sites exhibited different survival trajectories and different rates of change of associated bacterial communities to site-specific compositions. We discuss the variability and potential interaction of environmental conditions, survival and coral-associated bacterial communities, and hence the importance of integrating these factors into future planning, initiation, and monitoring of active reef management approaches to optimise success.
Site differences in out-plant survival
Reef rehabilitation practitioners face the challenge of highly variable survivorship of coral propagules with limited understanding of which factors primarily influence survival (e.g. Caribbean, Young et al. 2012; Lirman et al. 2014, and Great Barrier Reef, Suggett et al. 2020; Howlett et al. 2022). Such a core gap in knowledge represents a fundamental road-block to successfully scaling propagation and out-planting protocols (Ware et al. 2020; Hein et al. 2020; Boström-Einarsson et al. 2020). Differences in the survival of corals transplanted between environments during management interventions potentially stem from factors including site selection (e.g. prevailing environmental conditions and corallivore presence) (Pausch et al. 2018; Ware et al. 2020), coral-associated bacterial community composition (Moriarty et al. 2020), as well as coral species, coral size and attachment type (Yap 2004; Goergen et al. 2018; Munasik 2020). In our current study, we standardised coral species and attachment type, and fragment size to explore the potential role of coral-associated bacterial communities and environmental conditions between sites. At the start of the experiment (TF), we observed differences in flow, light attenuation and temperature across the study sites. Overall coral survival was generally higher at the site with historically the lowest recent impact (“Mojo”, Edmondson pers. obs.; see also Roper et al. 2022) that was initially characterised by higher flow rates, light attenuation and lower temperatures. Such factors have previously been described to promote resilience of corals at other sites (low temperature, Mediterranean—Rubio-Portillo et al. 2014; high flow, Palmyra Atoll—Rogers et al. 2016; low temperature and increased light attenuation/heterotrophy, Red Sea—Tremblay et al. 2016; increased light attenuation/heterotrophy, Caribbean, Indian and Pacific Ocean—Fox et al. 2018); however, it is unknown if initial environmental differences between sites were consistent throughout the experiment. Importantly, out-planting of Acropora spp. at Rayban has previously yielded 70–100% survival (mixed species assemblages after 3–7 months, Suggett et al. 2020; A. gemnifera, A. intermedia, A. spathulata after 11 months; Howlett et al. 2022) in contrast to the 0–35% survival after 12 months observed across plots within the same site in our current study. Further, similar bacterial community composition and low overall survival were observed at sites Rayban and Sandbox (0–43%) despite different initial environmental conditions (flow, temperature, and light attenuation), indicating the need for finer resolution spatiotemporal monitoring of environmental conditions and associated bacterial communities within sites as well as between sites with differential out-plant survival. Resolving the relationships between environmental conditions and bacterial community change in out-planted corals, and identifying differentiating factors between sites with low versus high out-plant survival will be instrumental in improving the success of restoration approaches through informed site selection.
Spatiotemporal dynamics of out-plant bacterial communities
Environmental heterogeneity is a suggested driver of host-microbiome composition (Dunphy et al. 2019) with nutrient gradients (Kelly et al. 2014), temperature variability (Littman et al. 2009; Santos et al. 2014; van Oppen and Blackall 2019), fish communities, (McDevitt-Irwin et al. 2017), pH and oxygen variability (Camp et al. 2020; Haydon et al. 2021)—all contributing and sometimes interacting synergistically towards reshaping coral-associated microbial communities (Maher et al. 2019). In our current study, we observed variability in the bacterial community composition of A. millepora after out-planting at three reef sites. This is consistent with previous observations for this genus, where transplanting corals between impacted sites and control sites (Ziegler et al. 2019), between mangrove and reef areas (Haydon et al. 2021) and specifically at Opal Reef previously during propagation and out-planting over 125 and 30 days (respectively) led to bacterial community changes (Maher et al. 2019; Strudwick et al. 2022). Interestingly, in our study, we observed bacterial community changes in out-planted coral at different times across sites. Whilst it remains unknown what specifically influences the rate of bacterial community change in corals over time, in other host–microbiome relationships, such as human–gut microbiome, responses to extreme changes in extrinsic factors (e.g. diet and exposure to foreign substances) can occur within days, whereas responses to mild changes in extrinsic factors occur within weeks to months (Uhr et al. 2019; Schlomann and Parthasarathy 2019). In the case of our study, the rate of bacterial community changes was variable between reef sites—7 days versus 1.5 months—suggesting the site where changes occurred more rapidly likely had the most different environmental conditions compared to the donor site. Interestingly, in relation to the environmental variables measured in this study, the site where bacterial communities changed within 7 days was more dissimilar to the donor site compared to the site where bacterial communities changed within 1.5 months. It is important to note there are likely variations in other environmental conditions (that we did not measure), such as pH (Zhang et al. 2015), algal exudates (Smith et al. 2006; Barott et al. 2012) and fish communities (Ezzat et al. 2019), between the donor and out-plant sites contributing to the site-specific rate of change and subsequent composition of coral-associated bacterial communities. It is integral to identify these contributing factors so restoration practitioners can estimate the likelihood of potential impacts to coral biology when transplanting coral fragments between sites with contrasting environmental conditions.
Potential links between microbial communities and out-plant survival
Associated bacterial communities are well known to play a role in resilience of the coral holobiont to environmental change (Reshef et al. 2006; Ainsworth and Gates 2016; Bourne et al. 2016; Glasl et al. 2016)—and hence the importance of considering the microbiome in reef management is integral (e.g. Voolstra et al. 2021). Certain bacteria have the capacity to play relevant roles in the coral holobiont functioning—and these may confer resilience, resistance or susceptibility to specific biotic and abiotic stressors—however, the mechanisms remain unresolved (Ben-Haim et al. 2003; Alagely et al. 2011; Bourne et al. 2016; Santos et al. 2015; Peixoto et al. 2017; Welsh et al. 2017; Rosado et al. 2019). Some species of Acropora thrive across a range of environmental conditions including those predicted to exist under future climate change scenarios found in mangrove lagoons (Camp et al. 2020) and can persist in these conditions in part due to specific bacterial community composition and/or changes in relative abundance of bacterial taxa in response to prevailing environmental conditions (Ziegler et al 2017, 2019). During our current study, distinct bacterial community restructuring was observed in out-plants that was consistent with our previous study where out-plants were monitored for only one month at Rayban (Strudwick et al. 2022). However, we observed site-specific post-out-planting microbiome changes accompanied by variable survival across sites with contrasting site topography and environmental conditions, suggesting the bacterial community changes observed were not necessarily a beneficial response to the novel environmental conditions of the out-planting site; rather, such changes potentially reflect loss of microbial taxa providing essential functions or dysbiosis (microbiome imbalance), thereby contributing to mortality (Egan and Gardiner 2016). Interestingly, increased heterogeneity of bacterial communities—previously suggested to indicate dysbiosis (Maher et al. 2019)—was paradoxically recorded at the site with the highest survival, and no signs of dysbiosis were recorded at the sites with the lowest survival. We therefore suggest, that observed initial differences in out-plant bacterial communities across sites may reflect a response to the novel local-scale environmental conditions at the out-planting site, with subsequent similarities across sites (in the bacterial communities) representing a successive period in which bacterial communities are primarily shaped by larger-scale environmental conditions across sites—such as global trends in temperature (Sharp et al. 2017; Cai et al. 2018)—and mortality is driven by other factors not measured in this study (e.g. corallivore presence) (Pausch et al. 2018).
Differences in bacterial community composition between the sites were explained by higher relative abundance of bacterial taxa (Ruegeria, Lactobacillus, Pseudovibrio and Erythrobacter) at the site with the highest survival (Mojo); these taxa have previously been linked to pathogen resistance (Karthikeyan and Santosh 2008—Lactobacillus; Kitamura et al. 2021; Miura et al. 2019; Rosado et al. 2019—Ruegeria; Raina et al. 2016; Pseudovibrio; Pereira et al. 2017—Erythrobacter). However, differences in bacterial taxa between sites were also explained by higher relative abundance of Endozoicomonas at the sites with lower survival (Sandbox and Rayban). Such an outcome is perhaps counter-intuitive where Endozoicomonas has also been linked to putatively beneficial functions in coral (Neave et al. 2016; Tandon et al. 2020) and are usually observed to decrease in abundance during stress (van Oppen and Blackall 2019), and hence contradictory to the higher mortality rates at these sites. Changes in the bacterial community structure of out-plants (at Mojo and Rayban) over time were also explained by fluctuations in the relative abundance of the core ASV from the Endozoicomonas genus and were consistent with previous observations of increased abundance of Endozoicomonas in February on the GBR (Epstein et al. 2019). We suggest similar changes in abundance of Endozoicomonas in out-plant bacterial communities across sites could indicate out-plant bacterial communities are not only influenced by local environmental conditions but also global trends in temperature and/or other environmental conditions. Putatively pathogenic bacteria were not recorded to proliferate in coral-associated bacterial communities at sites with lower out-plant survival. However, sampling dying coral where pathogens were likely to be present was not possible, as such we cannot confirm if a driver of out-plant mortality was microbial disease. Considering bacteria underpin the health and resilience of reef ecosystems (Reshef et al. 2006; Rosenberg et al. 2007; Ainsworth and Gates 2016; Bourne et al. 2016; van Oppen and Blackall 2019; van Oppen and Blackall 2019) and that bacterial communities associated with out-plants at sites of high mortality differed from out-plants at sites with low mortality, our study reinforces the importance of considering bacterial communities in the suite of factors influencing survival of out-planted corals. Future investigations into the role and source of specific bacterial taxa associated with out-planted corals at sites with high survival versus low survival will improve understanding of site selection for increased success.
Importance of integrating microbiome in planning, initiation and monitoring of future active reef management
For the first time, we have shown long-term site-specific changes in coral-associated bacterial communities of out-planted fragments alongside differences in out-plant survival and environmental conditions within a reef. Our findings suggest that contrasting environmental conditions between sites of the same reef could have an important role in survival of out-planted corals and likely influence speed of change and composition of coral-associated bacterial communities. These findings emphasise that improved understanding of the mechanisms through which environmental conditions impact coral-associated bacterial communities, and how this in turn effects coral host health, is essential to guide optimisation of restoration activities through improved site selection; in particular, in terms of out-planting site relative to nursery propagation site. Although we did not observe proliferation of putative pathogens at sites with lower coral out-plant survival, it is critical to note that increased abundance of microbes with the potential to incite pathogenesis have been observed at degraded sites (Haas et al. 2016; Silveira et al. 2017) and depending on the mechanisms of bacterial community change or the inherent microbiome variability of the host coral species, corals out-planted at these sites could remain vulnerable to pathogenesis (Thurber et al. 2009; Sato et al. 2016; Ziegler et al. 2019; Moriarty et al. 2020). On the contrary, coral species that have variable bacterial communities—such as Acropora spp.—may exhibit higher survival through bacterial community ‘conforming’ to the out-planting environment, compared to coral species that maintain stable bacterial communities potentially poorly suited to novel environments (Ziegler et al. 2019). Recent enthusiasm to use environmental DNA to improve reef monitoring methods (West et al. 2020; Richards et al. 2022) and probiotics during reef restoration (Peixoto et al. 2021) will have limited scope without thorough understanding of how microbial communities are influencing the health of rehabilitated reef ecosystems and how bacterial communities are influenced by active management processes. Therefore, it is critical to resolve the mechanisms of bacterial community change in out-planted corals, including through experiments involving systematic moderation of environmental conditions, to (i) clarify their role in out-plant mortality, (ii) identify whether findings hold across coral species and (iii) how the responses observed in this study translate across sites with varying degrees of degradation prior to expansion of activities into more degraded areas. Our results indicate that careful selection of appropriate out-planting sites has the potential to improve success of interventions, but it will only be possible to inform site selection by first thoroughly investigating the role of microorganisms in the survival/mortality of out-planted corals.
Data availability
Raw data files used for coral and environmental bacterial community analysis in this study have been deposited in FASTQ format in NCBI Sequence Read Archive (SRA) under Bioproject number PRJNA929655. All other data are provided as Electronic Supplementary Materials.
References
Ainsworth TD, Gates RD (2016) Corals’ microbial sentinels: the coral microbiome will be key to future reef health. Science 352(6293):1518–1519. https://doi.org/10.1126/SCIENCE.AAD9957
Alagely A, Krediet CJ, Ritchie KB (2011) Teplitski M (2011) Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J 5(10):1609–1620. https://doi.org/10.1038/ismej.2011.45
Barott KL, Rohwer FL (2012) Unseen players shape benthic competition on coral reefs. Trends Microbiol 20(12):621–628. https://doi.org/10.1016/j.tim.2012.08.004
Bayraktarov E, Saunders MI, Abdullah S, Mills M, Beher J, Possingham HP, Mumby PJ, Lovelock CE (2016) The cost and feasibility of marine coastal restoration. Ecol Appl 26(4):1055–1074. https://doi.org/10.1890/15-1077
Ben-Haim Y, Zicherman-Keren M, Rosenberg E (2003) Temperature-regulated bleaching and lysis of the coral Pocillopora damicomis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol 69(7):4236–4242. https://doi.org/10.1128/AEM.69.7.4236-4242.2003/ASSET/C3395145-6F34-4746-9EEB-38C4C31DFC78/ASSETS/GRAPHIC/AM0730017005.JPEG
Boström-Einarsson L, Ceccarelli D, Babcock RC, Bayraktarov E, Cook N, Harrison P, Hein M, Shaver E, Smith A, Stewart-Sinclair PJ, Vardi T, McLeod IM. (2018) Coral Restoration in a changing world A global synthesis of methods and techniques. Report. Reef and Rainforest Research Centre Ltd, Cairns, ISBN: 9781925514315. Sponsored by NESP Tropical Water Quality Hub. https://researchonline.jcu.edu.au/59790/1/59790_2019%20NESP%20Project%204.3%20Technical%20Report.pdf
Boström-Einarsson L, Babcock RC, Bayraktarov E, Ceccarelli D, Cook N, Ferse SCA, Hancock B, Harrison P, Hein M, Shaver E, Smith A, Suggett D, Stewart-Sinclair PJ, Vardi T, McLeod IM (2020) Coral restoration—a systematic review of current methods, successes, failures and future directions. PLoS ONE 15(1):e0226631. https://doi.org/10.1371/JOURNAL.PONE.0226631
Bourne DG, Morrow KM, Webster NS (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340. https://doi.org/10.1146/ANNUREV-MICRO-102215-095440
Cai L, Zhou G, Tong H, Tian RM, Zhang W, Ding W, Liu S, Huang H, Qian PY (2018) Season structures prokaryotic partners but not algal symbionts in subtropical hard corals. Appl Microbiol Biotechnol 102(11):4963–4973. https://doi.org/10.1007/S00253-018-8909-5/FIGURES/6
Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2 High-resolution sample inference from Illumina amplicon data. Nat Methods 13(7):581–583
Camp EF, Suggett DJ, Pogoreutz C, Nitschke MR, Houlbreque F, Hume BCC, Gardner SG, Zampighi M, Rodolfo-Metalpa R, Voolstra CR (2020) Corals exhibit distinct patterns of microbial reorganisation to thrive in an extreme inshore environment. Coral Reefs 39(3):701–716. https://doi.org/10.1007/s00338-019-01889-3
Casey JM, Connolly SR, Ainsworth TD (2015) Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci Rep 5(1):11903. https://doi.org/10.1038/srep11903
Crookshanks S (2008) High-Energy Sedimentary Processes in Kluane Lake, Yukon Territory. Doctoral dissertation, Queen’s University, Kingston, Ontario, Canada. https://www.collectionscanada.gc.ca/obj/s4/f2/dsk3/OKQ/TC-OKQ-1219.pdf. Accessed 5 Jan 2023
Damjanovic K, Blackall LL, Peplow LM, van Oppen MJH (2020) Assessment of bacterial community composition within and among Acropora loripes colonies in the wild and in captivity. Coral Reefs 39(5):1245–1255. https://doi.org/10.1007/s00338-020-01958-y
Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Thurber RV, Willis BL, Azam F, Knowlton N, Rohwer F (2008) Microbial ecology of four coral Atolls in the Northern Line Islands. PLoS ONE 3(2):e1584. https://doi.org/10.1371/JOURNAL.PONE.0001584
Duarte CM, Agusti S, Barbier E, Britten GL, Castilla JC, Gattuso JP, Fulweiler RW, Hughes TP, Knowlton N, Lovelock CE, Lotze HK, Predragovic M, Poloczanska E, Roberts C, Worm B (2020) Rebuilding marine life. Nature 580(7801):39–51. https://doi.org/10.1038/s41586-020-2146-7
Dunphy CM, Gouhier TC, Chu ND, Vollmer SV (2019) Structure and stability of the coral microbiome in space and time. Sci Rep 9(1):1–13. https://doi.org/10.1038/s41598-019-43268-6
Eakin CM, Sweatman HPA, Brainard RE (2019) The 2014–2017 global-scale coral bleaching event: insights and impacts. Coral Reefs 38(4):539–545. https://doi.org/10.1007/S00338-019-01844-2/METRICS
Eddy TD, Lam VWY, Reygondeau G, Cisneros-Montemayor AM, Greer K, Palomares MLD, Bruno JF, Ota Y, Cheung WWL (2021) Global decline in capacity of coral reefs to provide ecosystem services. One Earth 4(9):1278–1285. https://doi.org/10.1016/J.ONEEAR.2021.08.016
Egan S, Gardiner M (2016) Microbial dysbiosis: rethinking disease in marine ecosystems. Front Microbiol 7:991. https://doi.org/10.3389/fmicb.2016.00991
Epstein HE, Smith HA, Cantin NE, Mocellin VJL, Torda G, van Oppen MJH (2019) Temporal variation in the microbiome of Acropora coral species does not reflect seasonality. Front Microbiol 10:1775. https://doi.org/10.3389/fmicb.2019.01775
Ezzat L, Lamy T, Maher RL, Munsterman KS, Landfield K, Schmeltzer ER, Gaulke CA, Burkepile DE, Thurber RV (2019) Surgeonfish feces increase microbial opportunism in reef-building corals. Mar Ecol Prog Ser 631:81–97. https://doi.org/10.3354/MEPS13119
Glasl B, Herndl GJ, Frade PR (2016) The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J 10(9):2280–2292. https://doi.org/10.1038/ISMEJ.2016.9
Goergen EA, Gilliam DS (2018) Outplanting technique, host genotype, and site affect the initial success of outplanted Acropora cervicornis. PeerJ. https://doi.org/10.7717/PEERJ.4433/SUPP-2
Haas AF, Fairoz MFM, Kelly LW, Nelson CE, Dinsdale EA, Edwards RA, Giles S, Hatay M, Hisakawa N, Knowles B, Lim YW, Maughan H, Pantos O, Roach TNF, Sanchez SE, Silveira CB, Sandin S, Smith JE, Rohwer F (2016) Global microbialization of coral reefs. Nat Microbiol 1(6):1–7. https://doi.org/10.1038/nmicrobiol.2016.42
Haydon TD, Seymour JR, Raina JB, Edmondson J, Siboni N, Matthews JL, Camp EF, Suggett DJ (2021) Rapid shifts in bacterial communities and homogeneity of Symbiodiniaceae in colonies of Pocillopora acuta transplanted between reef and mangrove environments. Front Microbiol 12:3131. https://doi.org/10.3389/FMICB.2021.756091/BIBTEX
Hein MY, Beeden R, Birtles A, Gardiner NM, le Berre T, Levy J, Marshall N, Scott CM, Terry L, Willis BL (2020) Coral restoration effectiveness: Multiregional snapshots of the long-term responses of coral assemblages to restoration. Diversity 12(4):153. https://doi.org/10.3390/D12040153
Hein MY, Staub F, Banaszack A, Dallison T, Deri W, Grimsditch G, Jacob F, McLeod I, Mead D, Moore T, Newlands M, Shaver E, Thornton H, Vardi T, Winters RS, Zoccola D (2021) ICRI-Mapping the global funding landscape for coral reef restoration. Int Coral Reef Init 23:2–19
Howlett L, Camp EF, Edmondson J, Henderson N, Suggett DJ (2021) Coral growth, survivorship and return-on-effort within nurseries at high-value sites on the Great Barrier Reef. PLoS ONE 16(1):1–16. https://doi.org/10.1371/journal.pone.0244961
Howlett L, Camp EF, Edmondson J, Edmondson J, Agius T, Hosp R, Coulthard P, Edmondson S, Suggett DJ (2022) Adoption of coral propagation and out-planting via the tourism industry to advance site stewardship on the northern Great Barrier Reef. Ocean Coastal Manag. 225:106199. https://doi.org/10.1016/J.OCECOAMAN.2022.106199
Hughes TP, Barnes ML, Bellwood DR, Cinner JE, Cumming GS, Jackson JBC, Kleypas J, van de Leemput IA, Lough JM, Morrison TH, Palumbi SR, van Nes EH, Scheffer M (2017) Coral reefs in the Anthropocene. In Nature 546(7656):82–90. https://doi.org/10.1038/nature22901
Karthikeyan V, Santosh SW (2008) Isolation and partial characterization of bacteriocin produced from Lactobacillus plantarum. Afr J Microbiol Res 3(5):233–239. https://www.researchgate.net/publication/239923027
Kelly LW, Williams GJ, Barott KL, Carlson CA, Dinsdale EA, Edwards RA, Haas AF, Haynes M, Lim YW, McDole T, Nelson CE, Sala E, Sandin SA, Smith JE, Vermeij MJA, Youle M, Rohwer F (2014) Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proc Natl Acad Sci USA 111(28):10227–10232. https://doi.org/10.1073/PNAS.1403319111/-/DCSUPPLEMENTAL/PNAS.201403319SI.PDF
Kirk JT (1994) Light and photosynthesis in aquatic ecosystems. Cambridge University Press, Cambridge
Kitamura R, Miura N, Ito M, Takagi T, Yamashiro H, Nishikawa Y, Nishimura Y, Kobayashi K, Kataoka M (2021) Specific detection of coral-associated ruegeria, a potential probiotic bacterium corals subtropical seawater. Marine Biotechnol 23(4):576–589. https://doi.org/10.1007/S10126-021-10047-2/FIGURES/7
Kleypas J, Allemand D, Anthony K, Baker AC, Beck MW, Hale LZ, Hilmi N, Hoegh-Guldberg O, Hughes T, Kaufman L, Kayanne H, Magnan AK, Mclead E, Mumby P, Palumbi S, Richmond RH, Rinkevich B, Steneck RS, Voolstra CR, Wachenfeld D, Gattuso J (2021) Designing a blueprint for coral reef survival. Biol Conserv 257:109107. https://doi.org/10.1016/j.biocon.2021.109107
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41(1):1–11. https://doi.org/10.1093/nar/gks808
Lirman D, Schopmeyer S, Galvan V, Drury C, Baker AC, Baums IB (2014) Growth dynamics of the threatened Caribbean staghorn coral Acropora cervicornis: Influence of host genotype, symbiont identity, colony size, and environmental setting. PLoS ONE 9(9):e107253. https://doi.org/10.1371/JOURNAL.PONE.0107253
Littman R, Willis B, Pfeffer C, Bourne D (2009) Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol Ecol 68(2):152–163. https://doi.org/10.1111/j.1574-6941.2009.00666.x
Maher RL, Rice MM, McMinds R, Burkepile DE, Vega Thurber R (2019) Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci Rep. https://doi.org/10.1038/s41598-019-43274-8
Marchioro GM, Glasl B, Engelen AH, Serrão EA, Bourne DG, Webster NS, Frade PR (2020) Microbiome dynamics in the tissue and mucus of acroporid corals differ in relation to host and environmental parameters. PeerJ 8:1–26. https://doi.org/10.7717/peerj.9644
McAfee D, Costanza R, Connell SD (2021) Valuing marine restoration beyond the ‘too small and too expensive.’ Trends Ecol Evol 36(11):968–971. https://doi.org/10.1016/J.TREE.2021.08.002
McDevitt-Irwin JM, Baum JK, Garren M, Vega Thurber RL (2017) Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci 4(8):1–16. https://doi.org/10.3389/fmars.2017.00262
McKnight DT, Huerlimann R, Bower DS, Schwarzkopf L, Alford RA, Zenger KR (2019) Methods for normalizing microbiome data: an ecological perspective. Methods Ecol Evol 10(3):389–400. https://doi.org/10.1111/2041-210X.13115
McLeod IM, Hein MY, Babcock R, Bay L, Bourne DG, Cook N, Doropoulos C, Gibbs M, Harrison P, Lockie S, van Oppen MJH, Mattocks N, Page CA, Randall CJ, Smith A, Smith HA, Suggett DJ, Taylor B, Vella KJ, Wachenfeld D, Boström-Einarsson L (2022) Coral restoration and adaptation in Australia: the first five years. PLoS ONE 17(11):e0273325. https://doi.org/10.1371/JOURNAL.PONE.0273325
Miura N, Motone K, Takagi T, Aburaya S, Watanabe S, Aoki W, Ueda M (2019) Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Marine Biotechnol 21(1):1–8. https://doi.org/10.1007/S10126-018-9853-1/FIGURES/1
Moriarty T, Leggat W, Huggett MJ, Ainsworth TD (2020) Coral disease causes, consequences, and risk within coral restoration. Trends Microbiol 28(10):793–807. https://doi.org/10.1016/j.tim.2020.06.002
Munasik A (2020) Coral transplantation on a multilevel substrate of Artificial Patch Reefs: effect of fixing methods on the growth rate of two Acropora species. Biodiversitas. 21(5):1816–1822. https://doc-pak.undip.ac.id/6574/3/Riwayat_Korespondensi_Karil_GB_Munasik.pdf. Accessed 5 Jan 2023
Neave MJ, Rachmawati R, Xun L, Michell CT, Bourne DG, Apprill A, Voolstra CR (2016) Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J 11(1):186–200. https://doi.org/10.1038/ismej.2016.95
Pausch RE, Williams DE, Miller MW (2018) Impacts of fragment genotype, habitat, and size on outplanted elkhorn coral success under thermal stress. Mar Ecol Prog Ser 592:109–117. https://doi.org/10.3354/MEPS12488
Pedregosa, F., et al. "sklearn. model_selection. GridSearchCV." (2011).
Peixoto RS, Rosado PM, de Leite DCA, Rosado AS, Bourne DG (2017) Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Frontiers Microbiol. https://doi.org/10.3389/fmicb.2017.00341
Peixoto RS, Sweet M, Villela HDM, Cardoso P, Thomas T, Voolstra CR, Høj L, Bourne DG (2021) Coral probiotics: premise, promise. Prospects 9:265–288. https://doi.org/10.1146/ANNUREV-ANIMAL-090120-115444
Pereira LB, Palermo BRZ, Carlos C, Ottoboni LMM (2017) Diversity and antimicrobial activity of bacteria isolated from different Brazilian coral species. FEMS Microbiol Lett 364(16):164. https://doi.org/10.1093/FEMSLE/FNX164
Raina JB, Tapiolas D, Motti CA, Foret S, Seemann T, Tebben J, Willis BL, Bourne DG (2016) Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 16(8):e2275. https://doi.org/10.7717/peerj.2275
Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E (2006) The coral probiotic hypothesis. Environ Microbiol 8(12):2068–2073. https://doi.org/10.1111/j.1462-2920.2006.01148.x
Richards ZT, Stat M, Heydenrych M, Dibattista JD, Richards ZT, Stat M, Heydenrych M, Dibattista JD (2022) Environmental DNA for biodiversity monitoring of coral reefs. Coral Reefs of the World. 15:203–224. https://doi.org/10.1007/978-3-031-07055-6_13
Rogers J, Monismith S, Koweek D, Torres W, Dunbar R (2016) Thermodynamics and hydrodynamics in an atoll reef system and their influence on coral cover. Limnol Oceanogr 61(6):2191–2206. https://doi.org/10.1002/lno.10365
Roper CD, Camp EF, Edmondson J, Suggett DJ (2022) Combined impacts of natural recruitment and active propagation for coral population recovery on the Great Barrier Reef. Mar Ecol Prog Ser 700:95–109. https://doi.org/10.3354/MEPS14184
Rosado PM, Leite DCA, Duarte GAS, Chaloub RM, Jospin G, Nunes da Rocha U, Saraiva PJ, Dini-Andreote F, Eisen JA, Bourne DG, Peixoto RS (2019) Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J 13(4):921–936. https://doi.org/10.1038/s41396-018-0323-6
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5(5):355–362. https://doi.org/10.1038/nrmicro1635
Santos HF, Carmo FL, Duarte G, Dini-Andreote F, Castro CB, Rosado AS, van Elsas JD, Peixoto RS (2014) Climate change affects key nitrogen-fixing bacterial populations on coral reefs. ISME J 8(11):2272–2279. https://doi.org/10.1038/ismej.2014.70
Santos HF, Duarte GAS, Rachid CTDC, Chaloub RM, Calderon EN, Marangoni LFDB, Bianchini A, Nudi AH, do Carmo FL, Van Elsas JD, Rosado AS, Castro CBE, Peixoto RS (2015) Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci Rep 5(1):1–11. https://doi.org/10.1038/srep18268
Sato Y, Civiello M, Bell SC, Willis BL, Bourne DG (2016) Integrated approach to understanding the onset and pathogenesis of black band disease in corals. Environ Microbiol 18(3):752–765. https://doi.org/10.1111/1462-2920.13122/SUPPINFO
Schlomann BH, Parthasarathy R (2019) Timescales of gut microbiome dynamics. Curr Opin Microbiol 50:56–63. https://doi.org/10.1016/J.MIB.2019.09.011
Sharp KH, Pratte ZA, Kerwin AH, Rotjan RD, Stewart FJ (2017) Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata. Microbiome 5(1):120. https://doi.org/10.1186/S40168-017-0329-8/FIGURES/7
Shaver EC, McLeod E, Hein MY, Palumbi SR, Quigley K, Vardi T, Mumby PJ, Smith D, Montoya-Maya P, Muller EM, Banaszak AT, McLeod IM, Wachenfeld D (2022) A roadmap to integrating resilience into the practice of coral reef restoration. Glob Change Biol 28(16):4751–4764. https://doi.org/10.1111/GCB.16212
Silveira CB, Cavalcanti GS, Walter JM, Silva-Lima AW, Dinsdale EA, Bourne DG, Thompson CC, Thompson FL (2017) Microbial processes driving coral reef organic carbon flow. FEMS Microbiol Rev 41(4):575–595. https://doi.org/10.1093/FEMSRE/FUX018
Smith JE, Shaw M, Edwards RA, Obura D, Pantos O, Sala E, Sandin SA, Smriga S, Hatay M, Rohwer FL (2006) Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol Lett 9(7):835–845. https://doi.org/10.1111/J.1461-0248.2006.00937.X
Strudwick P, Seymour J, Camp EF, Edmondson J, Haydon T, Howlett L, Le Reun N, Siboni N, Suggett DJ (2022) Impacts of nursery-based propagation and out-planting on coral-associated bacterial communities. Coral Reefs 2021:1–18. https://doi.org/10.1007/S00338-021-02207-6
Suggett DJ, van Oppen MJH (2022) Horizon scan of rapidly advancing coral restoration approaches for 21st century reef management. Emerg Top Life Sci 6(1):125–136. https://doi.org/10.1042/ETLS20210240
Suggett DJ, Camp EF, Edmondson J, Boström-Einarsson L, Ramler V, Lohr K, Patterson JT (2019) Optimizing return-on-effort for coral nursery and outplanting practices to aid restoration of the Great Barrier Reef. Restor Ecol 27(3):683–693. https://doi.org/10.1111/rec.12916
Suggett DJ, Edmondson J, Howlett L, Camp EF (2020) Coralclip®: a low-cost solution for rapid and targeted out-planting of coral at scale. Restor Ecol 28(2):289–296. https://doi.org/10.1111/rec.13070
Sully S, Hodgson G, van Woesik R (2022) Present and future bright and dark spots for coral reefs through climate change. Glob Change Biol 28(15):4509–4522. https://doi.org/10.1111/GCB.16083
Tandon K, Lu CY, Chiang PW, Wada N, Yang SH, Chan YF, Chen PY, Chang HY, Chiou YJ, Chou MS, Chen WM, Tang SL (2020) Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J 14(5):1290–1303. https://doi.org/10.1038/s41396-020-0610-x
Thurber RV, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F (2009) Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11(8):2148–2163. https://doi.org/10.1111/j.1462-2920.2009.01935.x
Tremblay O, Gori A, Maguer J, Hoogenboom M, Ferrier-Pagès C (2016) Heterotrophy promotes the re-establishment of photosynthate translocation in a symbiotic coral after heat stress. Sci Rep 6(1):1–14. https://doi.org/10.1038/srep38112
Uhr GT, Dohnalová L, Thaiss CA (2019) The dimension of time in host-microbiome interactions. Msystems. https://doi.org/10.1128/MSYSTEMS.00216-18/ASSET/8B684660-8BF1-47D4-B7DF-8904471BAC54/ASSETS/GRAPHIC/MSYSTEMS.00216-18-F0001.JPEG
van Oppen MJH, Blackall LL (2019) Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 17(9):557–567. https://doi.org/10.1038/s41579-019-0223-4
van Woesik R, Ripple K, Miller SL (2018) Macroalgae reduces survival of nursery-reared Acropora corals in the Florida reef tract. Restor Ecol 26(3):563–569. https://doi.org/10.1111/REC.12590
van Woesik R, Banister RB, Bartels E, Gilliam DS, Goergen EA, Lustic C, Maxwell K, Moura A, Muller EM, Schopmeyer S, Winters RS, Lirman D (2021) Differential survival of nursery-reared Acropora cervicornis outplants along the Florida reef tract. Restor Ecol 29(1):e13302. https://doi.org/10.1111/REC.13302
Voolstra CR, Suggett DJ, Peixoto RS, Parkinson JE, Quigley KM, Silveira CB, Sweet M, Muller EM, Barshis DJ, Bourne DG, Aranda M (2021) Extending the natural adaptive capacity of coral holobionts. Nature Rev Earth Environ 2(11):747–762. https://doi.org/10.1038/s43017-021-00214-3
Ware M, Garfield EN, Nedimyer K, Levy J, Kaufman L, Precht W, Scott Winters R, Miller SL (2020) Survivorship and growth in staghorn coral (Acropora cervicornis) outplanting projects in the Florida Keys National Marine Sanctuary. PLoS ONE 15(5):e0231817. https://doi.org/10.1371/JOURNAL.PONE.0231817
Welsh RM, Rosales SM, Zaneveld JR, Payet JP, McMinds R, Hubbs SL, Thurber RLV (2017) Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ 5(5):e3315. https://doi.org/10.7717/peerj.3315
West KM, Stat M, Harvey ES, Skepper CL, DiBattista JD, Richards ZT, Travers MJ, Newman SJ, Bunce M (2020) eDNA metabarcoding survey reveals fine-scale coral reef community variation across a remote, tropical island ecosystem. Mol Ecol 29(6):1069–1086. https://doi.org/10.1111/MEC.15382
Weyrich LS, Farrer AG, Eisenhofer R, Arriola LA, Young J, Selway CA, Handsley-Davis M, Adler CJ, Breen J, Cooper A (2019) Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour 19(4):982–996. https://doi.org/10.1111/1755-0998.13011
Williams SL, Sur C, Janetski N, Hollarsmith JA, Rapi S, Barron L, Heatwole SJ, Yusuf AM, Yusuf S, Jompa J (2019) Large-scale coral reef rehabilitation after blast fishing in Indonesia. Restor Ecol 27(2):447–456
Yap HT (2004) Differential survival of coral transplants on various substrates under elevated water temperatures. Mar Pollut Bull 49(4):306–312. https://doi.org/10.1016/j.marpolbul.2004.02.017
Young CN, Schopmeyer SA, Lirman D (2012) A review of reef restoration and coral propagation using the threatened genus Acropora in the Caribbean and western Atlantic. Bull Mar Sci 88(4):1075–1098. https://doi.org/10.5343/BMS.2011.1143
Zhang YY, Ling J, Yang QS, Wang YS, Sun CC, Sun HY, Feng JB, Jiang YF, Zhang YZ, Wu ML, de Dong J (2015) The diversity of coral associated bacteria and the environmental factors affect their community variation. Ecotoxicology 24(7–8):1467–1477. https://doi.org/10.1007/S10646-015-1454-4/FIGURES/5
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun. https://doi.org/10.1038/ncomms14213
Ziegler M, Grupstra CGB, Barreto MM, Eaton M, BaOmar J, Zubier K, Al-Sofyani A, Turki AJ, Ormond R, Voolstra CR (2019) Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun. https://doi.org/10.1038/s41467-019-10969-5
Acknowledgements
The authors wish to express their gratitude to the Great Barrier Reef Marine Park Authority, whose support established the permit for the coral nurseries and out-planting at Opal Reef (G20/43740.1 and G21/45224.1 to EFC, DJS and JE), as well as staff from Wavelength Reef Cruises, who have continuously supported the project.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This research was supported by an Australian Government Research Training Program (RTP) Fee Offset Scholarship and Stipend to PS. Operations at Opal Reef—including site access—were supported by funding to the Coral Nurture Program from the Australian & Queensland Governments (“Solving the bottleneck of reef rehabilitation through boosting coral abundance: Miniaturising and mechanising coral out-planting” to DJS, EFC, JE). Contribution of EFC was through the University of Technology Sydney Chancellor’s Postdoctoral Research Fellowship and ARC Discovery Early Career Research Award (DE190100142).
Author information
Authors and Affiliations
Contributions
PS: Writing—original draft, formal analysis, investigation, conceptualisation, fieldwork—experiment initiation, sample collection, laboratory processing; DS and JS: writing—review & editing, supervision, conceptualisation; EC: writing—review & editing, supervision, conceptualisation, fieldwork—experiment initiation; JE: fieldwork—facilitation and coordination of field operations, experiment initiation, sample collection; CR and LH: Fieldwork—sample collection.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest for this submission.
Additional information
Responsible Editor: C.R. Voolstra.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Strudwick, P., Seymour, J., Camp, E.F. et al. Bacterial communities associated with corals out-planted on the Great Barrier Reef are inherently dynamic over space and time. Mar Biol 170, 85 (2023). https://doi.org/10.1007/s00227-023-04235-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-023-04235-y