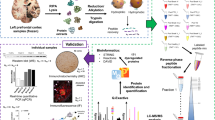
Abstract
Hepatic encephalopathy (HE) is a debilitating neurological disorder associated with liver failure and characterized by impaired brain function. Decade-long studies have led to significant advances in our understanding of HE; however, effective therapeutic management of HE is lacking, and HE continues to be a significant cause of morbidity and mortality in patients, underscoring the need for continued research into its pathophysiology and treatment. Accordingly, the present study provides a comprehensive overview aimed at elucidating the molecular underpinnings of HE and identifying potential therapeutic targets. A moderate-grade HE model was induced in rats using thioacetamide, which simulates the liver damage observed in patients, and its impact on cognitive function, neuronal arborization, and cellular morphology was also evaluated. We employed label-free LC–MS/MS proteomics to quantitatively profile hippocampal proteins to explore the molecular mechanism of HE pathogenesis; 2175 proteins were identified, 47 of which exhibited significant alterations in moderate-grade HE. The expression of several significantly upregulated proteins, such as FAK1, CD9 and Tspan2, was further validated at the transcript and protein levels, confirming the mass spectrometry results. These proteins have not been previously reported in HE. Utilizing Metascape, a tool for gene annotation and analysis, we further studied the biological pathways integral to brain function, including gliogenesis, the role of erythrocytes in maintaining blood–brain barrier integrity, the modulation of chemical synaptic transmission, astrocyte differentiation, the regulation of organ growth, the response to cAMP, myelination, and synaptic function, which were disrupted during HE. The STRING database further elucidated the protein‒protein interaction patterns among the differentially expressed proteins. This study provides novel insights into the molecular mechanisms driving HE and paves the way for identifying novel therapeutic targets for improved disease management.









Similar content being viewed by others
Data availability
The raw mass spectrometry data are available from the ProteomeXchange consortium and can be accessed via the dataset identifier PXD036723 (Username: reviewer_pxd036723@ebi.ac.uk Password: Jcvbq2v0). Supplementary figures and tables can be found in the shared supplementary files. Other data related to this work can be shared upon reasonable request to the corresponding author.
References
Amiri P, DeCastro J, Littig J et al (2021) Erythrocytes, a new contributor to age-associated loss of blood-brain barrier integrity. Adv Sci 8:2101912. https://doi.org/10.1002/advs.202101912
Anamika, Trigun SK (2021) Sirtuin-3 activation by honokiol restores mitochondrial dysfunction in the hippocampus of the hepatic encephalopathy rat model of ammonia neurotoxicity. Journal of Biochemical and Molecular Toxicology 35:e22735. https://doi.org/10.1002/jbt.22735
Anand KS, Dhikav V (2012) Hippocampus in health and disease: an overview. Ann Indian Acad Neurol 15:239–246. https://doi.org/10.4103/0972-2327.104323
Antunes M, Biala G (2012) The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process 13:93–110. https://doi.org/10.1007/s10339-011-0430-z
Banerjee SA, Hadjiargyrou M, Patterson PH (1997) An Antibody to the tetraspan membrane protein CD9 promotes neurite formation in a partially α3β1 integrin-dependent manner. J Neurosci 17:2756–2765. https://doi.org/10.1523/JNEUROSCI.17-08-02756.1997
Brunelli L, Campagna R, Airoldi L et al (2012) Exploratory investigation on nitro- and phospho-proteome cerebellum changes in hyperammonemia and hepatic encephalopathy rat models. Metab Brain Dis 27:37–49. https://doi.org/10.1007/s11011-011-9268-4
Buckingham JC, Flower RJ (2017) Chapter 25—annexin A1. In: Fink G (ed) Stress: neuroendocrinology and neurobiology. Academic Press, San Diego, pp 257–263
Bustin SA, Benes V, Garson JA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time pcr experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Butterworth RF (2003) Hepatic encephalopathy. Alcohol Res Health 27:240–246
Butterworth RF (2020) Hepatic encephalopathy. In: Irwin MA, Harvey JA, James LB, David EC, David AS, Snorri ST, Allan WW (eds) The liver: biology and pathobiology. John Wiley and Sons Ltd, pp 615–629
Chacón MR, Navarro AI, Cuesto G et al (2012) Focal adhesion kinase regulates actin nucleation and neuronal filopodia formation during axonal growth. Development 139:3200–3210. https://doi.org/10.1242/dev.080564
Chen R, Zhang Q, Yan Y et al (2021) Legumain knockout protects against Aβ1–42-induced AD-like cognitive deficits and synaptic plasticity dysfunction via inhibiting neuroinflammation without cleaving APP. Mol Neurobiol 58:1607–1620. https://doi.org/10.1007/s12035-020-02219-3
DeCarolis NA, Eisch AJ (2010) Hippocampal neurogenesis as a target for the treatment of mental illness: a critical evaluation. Neuropharmacology 58:884–893. https://doi.org/10.1016/j.neuropharm.2009.12.013
Deissler H, Blass-Kampmann S, Kindler-Röhrborn A et al (1996) Characterization of rat NCA/CD9 cell surface antigen and its expression by normal and malignant neural cells. J Neurosci Res 43:664–674
Deutsch EW, Csordas A, Sun Z et al (2017) The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res 45:D1100–D1106. https://doi.org/10.1093/nar/gkw936
Díaz-Hernandez M, del Puerto A, Díaz-Hernandez JI et al (2008) Inhibition of the ATP-gated P2X7 receptor promotes axonal growth and branching in cultured hippocampal neurons. J Cell Sci 121:3717–3728. https://doi.org/10.1242/jcs.034082
Dickson BJ (2002) Molecular mechanisms of axon guidance. Science 298:1959–1964. https://doi.org/10.1126/science.1072165
Doh-Ura K, Mekada E, Ogomori K, Iwaki T (2000) Enhanced CD9 expression in the mouse and human brains infected with transmissible spongiform encephalopathies. J Neuropathol Exp Neurol 59:774–785. https://doi.org/10.1093/jnen/59.9.774
Felipo V, Butterworth RF (2002) Neurobiology of ammonia. Prog Neurobiol 67:259–279. https://doi.org/10.1016/S0301-0082(02)00019-9
Ganaie IA, Malik MZ, Naqvi SH et al (2020) Differential levels of Alpha-1-inhibitor III, immunoglobulin heavy chain variable region, and hypertrophied skeletal muscle protein GTF3 in rat mammary tumorigenesis. Biochimie 174:57–68. https://doi.org/10.1016/j.biochi.2020.04.013
Grant SG, Karl KA, Kiebler MA, Kandel ER (1995) Focal adhesion kinase in the brain: novel subcellular localization and specific regulation by Fyn tyrosine kinase in mutant mice. Genes Dev 9:1909–1921. https://doi.org/10.1101/gad.9.15.1909
Hazell AS, Butterworth RF (1999) Hepatic encephalopathy: an update of pathophysiologic mechanisms. Proc Soc Exp Biol Med 222:99–112
Hitchcock IS, Skerry TM, Howard MR, Genever PG (2003) NMDA receptor–mediated regulation of human megakaryocytopoiesis. Blood 102:1254–1259. https://doi.org/10.1182/blood-2002-11-3553
Hořejši V, Vlček Č (1991) Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett 288:1–4. https://doi.org/10.1016/0014-5793(91)80988-F
Hwang I-H, Park J, Kim JM et al (2016) Tetraspanin-2 promotes glucotoxic apoptosis by regulating the JNK/β-catenin signaling pathway in human pancreatic β cells. FASEB J 30:3107–3116. https://doi.org/10.1096/fj.201600240RR
Iwasaki T, Takeda Y, Maruyama K et al (2013) Deletion of tetraspanin CD9 diminishes lymphangiogenesis in vivo and in vitro. J Biol Chem 288:2118–2131. https://doi.org/10.1074/jbc.M112.424291
Jaffe A, Lim JK, Jakab SS (2020) Pathophysiology of hepatic encephalopathy. Clin Liver Dis 24:175–188. https://doi.org/10.1016/j.cld.2020.01.002
Kagawa T, Mekada E, Shishido Y, Ikenaka K (1997) Immune system-related CD9 is expressed in mouse central nervous system myelin at a very late stage of myelination. J Neurosci Res 50:312–320. https://doi.org/10.1002/(SICI)1097-4547(19971015)50:2%3c312::AID-JNR19%3e3.0.CO;2-9
Khanna A, Anamika CS et al (2020) SIRT1 activation by resveratrol reverses atrophy of apical dendrites of hippocampal CA1 pyramidal neurons and neurobehavioral impairments in moderate grade hepatic encephalopathy rats. J Chem Neuroanat 106:101797. https://doi.org/10.1016/j.jchemneu.2020.101797
Kuleshova ID, Zaripov PI, Poluektov YM et al (2023) Changes in hemoglobin properties in complex with glutathione and after glutathionylation. Int J Mol Sci 24:13557. https://doi.org/10.3390/ijms241713557
Lee J-Y, Joo B, Nam JH et al (2018) An aqueous extract of herbal medicine ALWPs enhances cognitive performance and inhibits LPS-induced neuroinflammation via FAK/NF-κB signaling pathways. Front Aging Neurosci 10:269
Li Z, Wang X, Wang X et al (2023) Research progress on the role of extracellular vesicles in neurodegenerative diseases. Transl Neurodegener 12:43. https://doi.org/10.1186/s40035-023-00375-9
Lindsey ML, Mayr M, Gomes AV et al (2015) Transformative impact of proteomics on cardiovascular health and disease. Circulation 132:852–872. https://doi.org/10.1161/CIR.0000000000000226
Mondal P, Trigun SK (2014) Pannexin1 as a novel cerebral target in pathogenesis of hepatic encephalopathy. Metab Brain Dis 29:1007–1015. https://doi.org/10.1007/s11011-014-9556-x
Mondal P, Trigun SK (2015) Bacopa monnieri Extract (CDRI-08) modulates the NMDA receptor subunits and nNOS-apoptosis axis in cerebellum of hepatic encephalopathy rats. Evid Based Complement Altern Med 2015:e535013. https://doi.org/10.1155/2015/535013
Monje FJ, Kim E-J, Pollak DD et al (2012) Focal adhesion kinase regulates neuronal growth, synaptic plasticity and hippocampus-dependent spatial learning and memory. NSG 20:1–14. https://doi.org/10.1159/000330193
Nakamura Y, Iwamoto R, Mekada E (1996) Expression and distribution of CD9 in myelin of the central and peripheral nervous systems. Am J Pathol 149:575–583
Novikova G, Kapoor M, Tcw J et al (2021) Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat Commun 12:1610. https://doi.org/10.1038/s41467-021-21823-y
Owen JD, Ruest PJ, Fry DW, Hanks SK (1999) Induced focal adhesion kinase (FAK) expression in FAK-null cells enhances cell spreading and migration requiring both auto- and activation loop phosphorylation sites and inhibits adhesion-dependent tyrosine phosphorylation of Pyk2. Mol Cell Biol 19:4806–4818. https://doi.org/10.1128/MCB.19.7.4806
Palanisamy CP, Pei J, Alugoju P et al (2023) New strategies of neurodegenerative disease treatment with extracellular vesicles (EVs) derived from mesenchymal stem cells (MSCs). Theranostics 13:4138–4165. https://doi.org/10.7150/thno.83066
Park YK, Goda Y (2016) Integrins in synapse regulation. Nat Rev Neurosci 17:745–756. https://doi.org/10.1038/nrn.2016.138
Pierzchala K, Simicic D, Sienkiewicz A et al (2022) Central nervous system and systemic oxidative stress interplay with inflammation in a bile duct ligation rat model of type C hepatic encephalopathy. Free Radic Biol Med 178:295–307. https://doi.org/10.1016/j.freeradbiomed.2021.12.011
Podergajs N, Motaln H, Rajčević U et al (2015) Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget 7:593–609
Poordad FF (2007) Review article: the burden of hepatic encephalopathy. Aliment Pharmacol Ther 25:3–9. https://doi.org/10.1111/j.1746-6342.2006.03215.x
Provenzano PP, Keely PJ (2009) The role of focal adhesion kinase in tumor initiation and progression. Cell Adh Migr 3:347–350
Rama R, Derlig K, Vießmann N et al (2022) Simiate and the focal adhesion kinase FAK1 cooperate in the regulation of dendritogenesis. Sci Rep 12:11274. https://doi.org/10.1038/s41598-022-14460-y
Reynolds JL, Mahajan SD (2020) Transmigration of tetraspanin 2 (Tspan2) siRNA via microglia derived exosomes across the blood brain barrier modifies the production of immune mediators by microglia cells. J Neuroimmune Pharmacol 15:554–563. https://doi.org/10.1007/s11481-019-09895-6
Rico B, Beggs HE, Schahin-Reed D et al (2004) Control of axonal branching and synapse formation by focal adhesion kinase. Nat Neurosci 7:1059–1069. https://doi.org/10.1038/nn1317
Rocha-Perugini V, Sánchez-Madrid F, Martínez del Hoyo G (2016) Function and dynamics of tetraspanins during antigen recognition and immunological synapse formation. Front Immunol. https://doi.org/10.3389/fimmu.2015.00653
Rose CF, Amodio P, Bajaj JS et al (2020) Hepatic encephalopathy: novel insights into classification, pathophysiology and therapy. J Hepatol 73:1526–1547. https://doi.org/10.1016/j.jhep.2020.07.013
Sathyasaikumar KV, Swapna I, Reddy PVB et al (2007) Co-administration of C-phycocyanin ameliorates thioacetamide-induced hepatic encephalopathy in Wistar rats. J Neurol Sci 252:67–75. https://doi.org/10.1016/j.jns.2006.10.014
Schmidt C, Künemund V, Wintergerst ES et al (1996) CD9 of mouse brain is implicated in neurite outgrowth and cell migration in vitro and is associated with the α6/β1 integrin and the neural adhesion molecule L1. J Neurosci Res 43:12–31. https://doi.org/10.1002/jnr.490430103
Schrimpf A, Knappe O, Qvartskhava N et al (2022) Hyperammonemia-induced changes in the cerebral transcriptome and proteome. Anal Biochem 641:114548. https://doi.org/10.1016/j.ab.2022.114548
Sholl DA (1956) The organization of the cerebral cortex. John Wiley
Singh S, Koiri RK, Trigun SK (2008) Acute and chronic hyperammonemia modulate antioxidant enzymes differently in cerebral cortex and cerebellum. Neurochem Res 33:103–113. https://doi.org/10.1007/s11064-007-9422-x
Singh S, Trigun SK (2010) Activation of neuronal nitric oxide synthase in cerebellum of chronic hepatic encephalopathy rats is associated with up-regulation of NADPH-producing pathway. Cerebellum 9:384–397. https://doi.org/10.1007/s12311-010-0172-y
Singh S, Trigun SK (2014) Low grade cirrhosis induces cognitive impairment and motor dysfunction in rats: could be a model for minimal hepatic encephalopathy. Neurosci Lett 559:136–140. https://doi.org/10.1016/j.neulet.2013.11.058
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Tan X, Yan Y, Song B et al (2023) Focal adhesion kinase: from biological functions to therapeutic strategies. Exp Hematol Oncol 12:83. https://doi.org/10.1186/s40164-023-00446-7
Terada N, Baracskay K, Kinter M et al (2002) The tetraspanin protein, CD9, is expressed by progenitor cells committed to oligodendrogenesis and is linked to β1 integrin, CD81, and Tspan-2. Glia 40:350–359. https://doi.org/10.1002/glia.10134
Termini CM, Gillette JM (2017) Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol. https://doi.org/10.3389/fcell.2017.00034
Titus ADJ, Shankaranarayana Rao BS, Harsha HN et al (2007) Hypobaric hypoxia-induced dendritic atrophy of hippocampal neurons is associated with cognitive impairment in adult rats. Neuroscience 145:265–278. https://doi.org/10.1016/j.neuroscience.2006.11.037
Vizcaíno JA, Deutsch EW, Wang R et al (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32:223–226. https://doi.org/10.1038/nbt.2839
Yaseen IH, Monk PN, Partridge LJ (2017) Tspan2: a tetraspanin protein involved in oligodendrogenesis and cancer metastasis. Biochem Soc Trans 45:465–475. https://doi.org/10.1042/BST20160022
Zacharias HD, Zacharias AP, Gluud LL, Morgan MY (2019) Pharmacotherapies that specifically target ammonia for the prevention and treatment of hepatic encephalopathy in adults with cirrhosis. Cochrane Database Syst Rev. https://doi.org/10.1002/14651858.CD012334.pub2
Zaqout S, Kaindl AM (2016) Golgi-Cox staining step by step. Front Neuroanat. https://doi.org/10.3389/fnana.2016.00038
Zhang Z, Song M, Liu X et al (2015) Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer’s disease. Nat Commun 6:8762. https://doi.org/10.1038/ncomms9762
Zhou Y, Zhou B, Pache L et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10:1523. https://doi.org/10.1038/s41467-019-09234-6
Acknowledgements
The authors acknowledge Prof. SK Trigun Laboratory facilities, Department of Zoology, BHU, Prof Sanjeeva Srivastava, BSBE, and IIT Bombay for the MASSFIITB facility (BT/PR13114/INF/22/206/2015), the Central Discovery Centre, BHU for Trinocular Research Microscope and the Interdisciplinary School of Life Sciences, BHU for the qPCR facility. SKP thanks the Science and Engineering Research Board, Government of India, for the project assistance fellowship and the Indian Council of Medical Research for a senior research fellowship. We also thank the University Grants Commission, India, for providing a Junior Research Fellowship to VVS.
Funding
This work was financially supported by an ECRA grant from the Science and Engineering Research Board (ECR/2017/001018), the Government of India, a Start Up Grant (Dev. Scheme No. 5007), and a Research Grant for Faculty (IoE Dev. Scheme-6031) from Banaras Hindu University to PA.
Author information
Authors and Affiliations
Contributions
Shambhu Kumar Prasad: investigation, formal analysis, validation, visualization, writing-original draft. Vishal Vikram Singh: investigation, writing-review. Arup Acharjee: conceptualization, formal analysis, writing-review, and editing. Papia Acharjee: conceptualization, funding acquisition, formal analysis, supervision, writing-review, and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This study was approved by the Institutional Animal Ethical Committee (IAEC), Banaras Hindu University, Varanasi, India.
Consent to publication
All the authors reviewed the manuscript and agreed to publish this article.
Additional information
Communicated by Sreedharan Sajikumar.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Prasad, S.K., Singh, V.V., Acharjee, A. et al. Elucidating hippocampal proteome dynamics in moderate hepatic encephalopathy rats: insights from high-resolution mass spectrometry. Exp Brain Res (2024). https://doi.org/10.1007/s00221-024-06853-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00221-024-06853-4