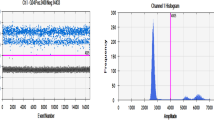
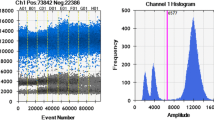
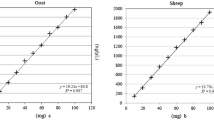
Abstract
Digital PCR methods were recently introduced in food analysis. To run these methods within the scope of ISO17025, they have to be validated. Although several guidelines are available, each laboratory has to implement these guidelines in an adapted validation scheme. We present here one possible implementation. We chose 13 GMO traits which were predominantly detected in the past. The results show that in the range of 1% GMO content, the digital PCR has a little superior performance compared to real time PCR. In the range of the detection limit, measurement uncertainty remains comparable to real time PCR. During validation, a conversion factor was determined for each trait suggesting that the calculation from % copies/copies to % weight/weight may be possible. This shows that GMO contents can be measured without the use of reference material by the validation described here and determination of a conversion factor, which is a great improvement in terms of expense and storage capacity.
Similar content being viewed by others
References
Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA (2011) Evaluation of digital PCR for absolute DNA quantification. Anal Chem 83(17):6474–6484
Pinheira LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emsli KR (2012) Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84:1003–1011
Gürtler P, Gerdes L, (2014) Digitale polymerasekettenreaktion, ddPCR; BIOspektrum | 06.14 | 20. Jahrgang, p. 632–635
Köppel René, Bucher Thomas (2015) Rapid establishment of droplet digital PCR for quantitative GMO analysis. Eur Food Res Technol 241(3):427–439
Dobnik D, Spilsberg B, Bogožalec Košir A, Holst-Jensen A, Žel J (2015) Multiplex quantification of 12 European union authorized genetically modified maize lines with droplet digital polymerase chain reaction. Anal Chem 87(16):8218–8226
Gerdes L, Iwobi A, Busch U, Pecoraro S (2016) Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomol Detect Quantif 7:9–20
Corbisier P, Bhat S, Partis L, Rui V, Xie D, Emslie KR (2010) Absolute quantification of genetically modified MON810 maize (Zea mays L.) by digital polymerase chain reaction. Anal Bioanal Chem 396(6):2143–2150
Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Rufa IK, Benjamin J, Hindson R, Vessella L, Tewari M (2013) Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10:1003–1005
Urquiza MP, Silva AA (2014) Copy number ratios determined by two digital polymerase chain reaction systems in genetically modified grains. Metrologia 51(1):61–66
European Union Reference Laboratory for GM Food and Feed (2008) Definition of minimum Performance requirements for analytical methods of GMO testing. http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf Accessed 29 Jan 2013
S. Trapman, Burns M, Broll H, Macarthur R, Wood R, Zel J (2009) Guidance document on measurement uncertainty for GMO testing laboratories. In: European Commission—Joint Research Centre—Institute for Reference Materials and Measurements, vol 1111, pp 1–41 (ISBN: 978-92-79-11228-7; p18, Equation 10)
Broedersa S, Huberb I, Grohmannc L, Berbend G, Tavernierse I, Mazzaraf M, Roosensa N, Morisset D (2014) Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci Technol 37:115e126
Scholdberg TA, Norden TD, Nelson D, Jenkins RG (2009) Evaluating precision and accuracy when quantifying different endogenous control, reference genes in maize using real-time PCR. J Agric Food Chem 57(7):2903
Zhang Nina Papazova David, Gruden Kristina, Vojvoda Jana, Yang Litao, Gašparič Meti Buh, Blejec Andrej, Fouilloux Stephane, De Loose Marc, Taverniers Isabel (2010) Evaluation of the reliability of maize reference assays for GMO quantification. Anal Bioanal Chem 396(6):2189–2201
Schaldberg TA, Norden TD, Nelson D, Jenkins GR (2009) Evaluating precision and accuracy when quantifying different endogenous control reference genes in maize using real time PCR. J Agric Food Chem 57:2903–2911
Köppel R, Dvorak V, Zimmerli F, Breitenmoser A, Eugster A, Waiblinger HU (2010) Two tetraplex real-time PCR for the detection and quantification of DNA from eight allergens in food. Eur Food Res Technol 230:367–374
Rönning SB (2003) Event specific real-time quantitative PCR for genetically modified Bt11 maize (Zea mays). Eur Food Res Technol 216:347–354
Nielsen Christer R, Berdal Knut G, Holst-Jensen Arne (2004) Charcterisation of the 5’ integration site and development of an event-specific real-time PCR assay for NK603 maize from a low starting copy number. Eur Food Res Technol 219:421–427
Lee S-H, Yi B-Y, Kim S-J (2009) Event-specific analytical methods for biotech maize MIR 604 and DAS-59122-7. J Sci Food Agric 89(15):2616–2624
Kim J-H, Kim H-Y (2009) Event-specific detection methods for genetically modified maize MIR604 using real-time PCR. Food Sci Biotechnol 18(5):1118–1123
La Paz J-L, Garca-Muniz N, Nadal A, Esteve T, Puigdomnech P, Pla M (2006) Interlaboratory transfer of a real-time polymerase chain reaction assay for quantitative detection of genetically modified maize event TC-1507. J AOAC Int 89(5):1347–1352
Köppel R, Bucher T, Frei A, Waiblinger H-U (2015) Droplet digital PCR versus multiplex real-time PCR Method for the detection and quantification of DNA from the four transgenic soy traits Mon87769, Mon87708, MON87705, FG72 and lectin. Eur Food Res Technol 241(4):521–527
Corbisier P, Barbante A, Berben G, Broothaerts W, De Loose M, Emons H, Georgieva TZ, Lievens A, Mazzara M, Papazova N, Perri E, Sowa S, Stebih D, Terzi V, Trapmann S (2017) Recommendation for the unit of measurement and the measuring system to report traceable and comparable results expressing GM content in accordance with EU legislation. Publ Office Eur Union. doi:10.2760/177516
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Rights and permissions
About this article
Cite this article
Köppel, R., Bucher, T., Bär, D. et al. Validation of 13 duplex droplet digital PCR systems for quantitative GMO analysis of most prevalent GMO traits. Eur Food Res Technol 244, 313–321 (2018). https://doi.org/10.1007/s00217-017-2957-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-017-2957-4