Abstract
Ultrasensitive detection of biomarkers is of paramount importance in various fields. Superior to the conventional ensemble measurement-based assays, single-entity assays, especially single-entity detection-based digital assays, not only can reach ultrahigh sensitivity, but also possess the potential to examine the heterogeneities among the individual target molecules within a population. In this review, we summarized the current biomolecular analysis methods that based on optical counting and imaging of the micro/nano-sized single entities that act as the individual reactors (e.g., micro-/nanoparticles, microemulsions, and microwells). We categorize the corresponding techniques as analog and digital single-entity assays and provide detailed information such as the design principles, the analytical performance, and their implementation in biomarker analysis in this work. We have also set critical comments on each technique from these aspects. At last, we reflect on the advantages and limitations of the optical single-entity counting and imaging methods for biomolecular assay and highlight future opportunities in this field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The high-sensitive detection of biomarkers is of great significance in biology, environmental science and engineering, food and agriculture industry, public health, and drug development. Among the various detection technologies, single-entity analysis provides a new breakthrough in the field of biomarker detection [1]. It is because different from the conventional ensemble measurements, in single-entity assays, each entity acting as the separate microreactor (e.g., micro-/nanoparticles, microemulsions, and microwells) to reflect the target content can be interrogated individually to provide dramatically improved detection sensitivity, which can even differentiate the heterogeneities among the individual target molecules within a population [2, 3]. Therefore, tremendous efforts have been devoted to the single-entity analysis, which mainly focus on two key aspects. In the first place, how to convert the target molecules’ concentration information to readable signals on the entities? Then, what are the appropriate ways to acquire the target-induced signals loaded on the single entities?
In this review, the biomolecular analysis methods based on counting and imaging of micro/nano single entities acting as the reactors are overviewed. On the one hand, considering the signal conversion mechanism, the single-entity assays are classified as analog assays and digital assays (Fig. 1). Specifically, in the analog assays, the targets are evenly distributed on the entities through Gaussian distribution, and there would be a certain correlation between the target biomolecule concentration and the mean signal intensity of all the interrogated entities [4], whereas in the digital assays, the number of the entities employed is typically far more than that of the target molecules [5,6,7]. In this way, one entity associates with only one or none target molecule following Poisson distribution, in which the individual entities loaded with one target molecule are recorded as “1” (positive) and those without target loaded are recorded as “0” (negative) after appropriate signal transduction. Therefore, the number of targets is directly reflected by the number of positive/negative entities. On the other hand, considering the majority of the existing single-entity assays, particularly the digital ones, mainly rely on the fluorescence signal readout pathway. Therefore, optical methods, especially fluorescence signals and some kinds of scattering signals would be the focus of this review due to their convenience and prevalence in single-entity counting and imaging. In this review, for convenient description, the “counting and imaging” is abbreviated as “counting” in the following description.
Diagram of the analog and digital optical counting-based single-entity assays. Top panel: analog measurements provide target concentration via fluorescence or other optical signal intensity variation. Bottom panel: digital measurements are independent of signal intensity variation, simply rely on counting the number of target-associated positive entities above a pre-set threshold
Based on the comprehensive introduction of the current progress in single entity-based biomolecular assays, the pros and cons of each method in this critical review are discussed. Moreover, in the last section, we point out several promising directions that may lead the future trends in single entity-based biomarker detection toward clinical diagnosis.
Single-entity optical counting-based analog bioassays
In a typical optical counting-based single-entity analog bioassay, the target molecules will induce the optical signal change on single entities. Generally, the optical signals of individual entities are all collected, while the mean intensity of the whole entity population is used to quantify the target dosage. The entities as reactors are in micro or nanoscale, which can be microbeads (MBs), micro nucleic acid amplicons, QDs, gold nanoparticles (AuNPs), magnetic nanoparticles (MNPs), etc. In this section, we will introduce the optical counting assays by classifying the reaction carries as micro entities and nano entities.
Optical counting of micro entities for analog bioassay
Entities in micron scale are popular reactors for optical counting assays because they can be easily detected with conventional microscope and flow cytometer (FCM) [8, 9]. Among them, Luminex technology is one of the most classical MB-based bioassays [10,11,12]. By tuning the ratio of different fluorescent dyes for encoding the MBs that correspond to specific type of targets, it permits the simultaneous labeling of a library of biomarkers. Consequently, the multiplexed detection of targets can be achieved by decoding the fluorescence color information and intensities on the MBs one by one in a liquid flow. For example, as shown in Fig. 2a, Li et al. designed a Luminex system with stem-loop-structured probes on the MBs for the detection of multiple microRNA (miRNA) targets [11]. In the initial stage, the probes were in a closed stem conformation, sterically shielding the biotin label from being accessible to the fluorescence reporter. After hybridizing with target miRNAs, the probes underwent a conformational switch, restoring accessibility of the biotin to streptavidin-phycoerythrin (STV-PE). In this way, the quantitative and qualitative assays of multiple miRNA targets can be achieved simultaneously by reading out the fluorescence code of the MB and the signal of PE.
a Schematic illustration of Luminex system for multiplex detection of miRNAs based on conformational switch of stem-loop probe upon target hybridization. Reprinted with permission of [11]. Copyright 2014, American Chemical Society. b Schematic illustration of the FCM strategy for the sensitive detection of miRNA based on the combining the cycling CNAL system and on-MB TdT extension. Reprinted with permission of [17]. Copyright 2018, American Chemical Society. c Schematic illustration of the MB-assisted TdT-FCM system for the detection of protein biomarkers. Reprinted with permission of [18]. Copyright 2018, Royal Society of Chemistry. d Schematic diagram of multiple nucleic acid target detection based on the single-miRNA initiated RCA. Reprinted with permission of [19]. Copyright 2020, American Chemical Society
Although the Luminex system enables multiplexed analysis of biomarkers, it requires the specialized instruments with complex optical path to discriminate the MBs encoded with different dyes. Therefore, exploiting the strategies that rely on the conventional FCM that widely equipped in routine research laboratories is important to the popularization of single entity-based assays. In addition, besides the multiplex analysis, the ability of detecting ultralow content of biomarkers is another key in clinical diagnosis. Therefore, various nucleic acid amplification strategies have been introduced into the micro entity-based assays depending on conventional FCM for the sensitive detection of biomarkers [13,14,15,16,17,18]. For example, for the high-sensitive detection of miRNA, Fan et al. integrated a two-step signal amplification with the single-entity analog assay, which includes the target-templated click nucleic acid ligation (CNAL) as the first step and the on-MB terminal deoxynucleotidyl transferase (TdT) enzymatic DNA polymerization as the second step (Fig. 2b) [17]. At first, the miRNA target initiated the cycling CNAL reaction in the solution to generate a number of ligated ssDNA with 5′-biotin and 3′-OH termini. After binding the ligated ssDNA on the magnetic MBs via the biotin-STV conjugation, the 3′-OH termini of the ssDNA molecules were recognized by TdT to initiate the template-free DNA polymerization and fluorescent ssDNA accumulation. Finally, FCM was employed to investigate the fluorescent signal of MBs one by one to reflect the concentration of miRNA. Furtherly, Zhu et al. reported an on-MB TdT immunoassay by employing the AuNPs co-functionalized with detection antibody and a 3′-OH ssDNA as probes (Fig. 2c) [18]. In the presence of the target protein, the AuNPs loaded with 3′-OH termini DNA molecules were combined with the capture antibody-coated MBs via the sandwich-type immunoreaction. TdT enzyme was then added to initiate the efficient extension reaction to yield long ssDNA molecules on the MBs to capture the fluorescent probes by hybridization. Finally, the amount of the target protein was evaluated by interrogating the fluorescent signals of the MB populations with FCM. This method could not only achieve the highly sensitive protein biomarker analysis, but also accomplished simultaneous analysis of multiple biomarkers based on the size discrimination capability of FCM. In addition to MBs, nucleic acid amplicons can also be used as the single entities for high-sensitive biomarker analysis. For example, as shown in Fig. 2d, Smith et al. reported a multiple nucleic acid target detection method based on the single-miRNA initiated rolling chain amplification (RCA) [19]. In their design, each miRNA was extended from its 3′ end by RCA to form miRNA amplicons. After fluorescently labeled with dye-conjugated ssDNA probes through hybridization, various micron-scale miRNA amplicons were analyzed with FCM. In this way, sensitive miRNA quantification was achieved with a detection limit of 47 fM, which was 2 orders of magnitude lower than the conventional qRT-PCR.
Compared with using large number of entities, the sensing performance will be highly improved if only one entity is employed as the reactor because the target-induced signals will be highly concentrated to allow for much enhanced sensitivity. Considering this, our group developed a series of single MB-based fluorescence imaging methods for the analysis of biomarkers [20,21,22]. For instance, Zhang et al. reported a single MB-based fluorescence imaging strategy for the detection of protein kinase (PK) activity in single cell lysates (Fig. 3a) [20]. The single MB used in this work was functionalized with rare earth (RE) ions via the interaction between RE ions and the phosphate backbone of ssDNA immobilized on the MB. In the presence of PKs, the fluorescent peptides could be phosphorylated and bound with RE ions on the MB. Finally, kinase activity was determined by reading the fluorescence signal on the single MB with a confocal fluorescence microscope. In another work, as shown in Fig. 3b, Chen et al. proposed an ultrasensitive immunoassay that could high efficiently enrich fluorophores on only a single MB [22]. In this assay, the detective antibody and biotinylated ssDNA were co-functionalized on the AuNPs. After the sandwiched immunoassay, large amounts of biotin groups on the AuNPs were brought on the single MB, furtherly resulting in the enrichment of STV-poly HRP. Then, in the presence of H2O2, the poly HRP catalyzed highly efficient tyramide signal amplification (TSA), covalently accumulating numerous biotin-tyramide molecules and finally loading a lot of STV-conjugated Alexa-Fluor 546 on the MB. Therefore, the antigen target was quantified by monitoring the fluorescence signal enriched in the tiny region of the MB surface with a confocal fluorescence microscope.
Single MB-based optical imaging analog bioassays. a Illustration of RE-functionalized single MB for capturing PKA-induced phosphopeptides toward PKA activity determination. Reprinted with permission of [20]. Copyright 2015, Wiley-VCH Verlag GmbH & Co. KGaA. b Schematic illustration of the ultrasensitive immunoassay by combining the AuNPs amplification and TSA on a single MB. Reprinted with permission of [22]. Copyright 2022, Royal Society of Chemistry. c Schematic interpretation of the single MB-based immunoassay via a plasmon-enhanced SERS mapping strategy. Reprinted with permission of [23]. Copyright 2020, American Chemical Society. d Scheme of the multiplexed miRNA assay on a single plasmonic MB. Reprinted with permission of [24]. Copyright 2021, American Chemical Society
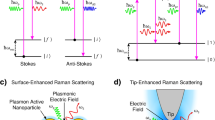
Surface-enhanced Raman scattering (SERS) is endowed with a collection of advantages, including the capability to provide the intrinsic molecular fingerprint information in biological systems, the photostability that allows long-term monitoring, the adaptability to both liquid-form and solid-form samples, and the narrow bandwidth of the characteristic peaks that enables multiplexed detection. Therefore, the mapping of SERS signal on an MB has emerged to be a powerful tool for bioassay. For example, Lu et al. have developed AuNP-AuNP bi-layer structure on a single MB for the multiplexed assay of protein and miRNA targets [23, 24]. As shown in Fig. 3c and d, a plasmonic AuNP layer carrying capture probes was pre-modified on the surface of the single MB. The target protein or miRNA will specifically introduce the SERS tag AuNPs to the MB surface via immunoreactions (sandwich-type antibody-antigen reaction or S9.6-DNA/RNA hybrid recognition), forming AuNP-AuNP gap structures to greatly enhance the SERS intensity and improve the detection sensitivity. By using a Raman scanning confocal microscope, the SERS signals concentrated on the MB surface can be resolved to reflect the target concentration. The single MB SERS assay can achieve high sensitivity to detect PSA protein and sub-pM miRNA. Moreover, by adopting various capture probe-SERS tag AuNP combinations, multiplexed assay can be facilely achieved. Besides the advantage of concentrating the SERS tags on a limited zone around the surface of only a single MB to improve the detection sensitivity, superior to the single-point measurements, SERS mapping throughout the MB surface is adopted for signal readout, providing per-pixel mean SERS intensity on the MB with greatly improved reproducibility. The single-MB mapping fashion will effectively suppress the signal fluctuation that generally exists in the conventional single-point measurement-based SERS methods.
Entities in micron scale are easily operated and suitable for the high-throughput detection instruments, which have been employed as efficient reaction carriers in the optical counting and imaging-based bioassays. However, they have relatively small surface-to-volume ratio. In this consideration, abundant number of reporter molecules should be enriched on the micro entities to make the signal detectable, to some extent limiting the detection sensitivity.
Optical counting of nano entities for analog bioassay
Compared with the micro entities, the ones in nanoscale have a smaller diameter and much larger surface-to-volume ratio to provide more surface area for specific reactions, which have greater potential to improve detection sensitivity. Therefore, nano entities have also been regarded as critical reactors in optical counting assay of biomarkers [25]. Among them, the ones based on quantum dots (QDs) show great promise. On the one hand, the large surface area of QDs facilitates the covalent conjugation of various biorecognition molecules for the preparation of fluorescent probes. On the other hand, QDs can act as fluorescence resonance energy transfer (FRET) donors or acceptors to construct various biosensors for the detection of biomarkers. Based on this, Zhang’s group developed a series of single-QD-based FRET biosensors [26,27,28]. For example, they proposed a single QD-mediated FRET nanosensor by employing multiple primer-generation RCA for the sensitive detection of single-nucleotide polymorphisms (SNPs) in cancer cells (Fig. 4a) [27]. With the assistance of nicking endonuclease, the presence of a mutant target induced cyclic multiple primer RCA, resulting in the exponential amplification and generation of large numbers of linker probes. The linker probes hybridized with Cy5-labeled reporter probes and the biotinylated capture probes and then bound on the QDs, leading to efficient FRET from the QDs to Cy5. By counting the FRET signal with total internal reflection fluorescent microscope (TIRFM), this method achieved a large dynamic range of 8 orders of magnitude and a detection limit of 0.0541 aM in SNP detection.
Single nano entity-based optical counting analog bioassays. a Principal illustration of the single QD-mediated FRET nanosensor for SNP assay. Reprinted with permission of [27]. Copyright 2021, American Chemical Society. b Illustration of the TIRFM assay for exosomes in plasma. Reprinted with permission of [29]. Copyright 2019, American Chemical Society. c Schematic illustration of the click chemical ligation-based on-MNP system for miRNA analysis. Reprinted with permission of [32]. Copyright 2018, American Chemical Society. d Schematic diagram of high-sensitivity detection of miRNA with DFM. Reprinted with permission of [36]. Copyright 2020, American Chemical Society
Even though the QD-based FRET strategy is free of washing and separation steps, efficient FRET requires the careful consideration of the distance and the angle between the donor and the acceptor, limiting its practical applications. Different from the FRET-based assays, many researches employed the nano entities as the carriers to enrich the optical signals. Typically, He et al. described an ultrasensitive exosome assay by enriching fluorescence signal on the membrane of exosomes (Fig. 4b) [29]. This assay employed specific aptamer probes (AAP) to capture target exosomes on a piece of glass-coverslip. The bound AAPs triggered the in situ hybridization chain reaction (HCR) to enrich numerous activated fluorescence molecules around exosomes. The fluorescence signal was then detected under a TIRFM system for quantitative analysis. This assay was successfully used for quantifying tumor exosomes in plasma and monitoring tumor progression and early responses to therapy.
Nano entities are generally not easy to be separated from the redundant probes due to their small size. To address this, many groups employed the MNPs in the optical imaging assays to simplify the separation procedure and eliminate the background signal [30,31,32,33]. For instance, Qi et al. reported a simple and highly sensitive enzyme-free miRNA assay by integrating cycling CNAL with TIRFM detection [32]. With the mediation of target miRNA, a lot of ligated products labeled with 5′-AlexaFluor488 and 3′-biotin could be produced by the click chemistry reaction (Fig. 4c). Then, the fluorescently ligated products were immobilized on the MNPs via STV-biotin conjugation. After magnetic separation, TIRFM imaging of the MNPs carrying fluorescent signals was carried out without interference from the free fluorescent probes. As a result, as low as 50 fM miRNA target could be detected with the ability to distinguish single base variation.
In addition to QDs and MNPs, AuNPs are another group nano entities for optical counting-based assays. AuNPs present a substantial molecular extinction cross-section, resonance Rayleigh scattering efficiency, and enhanced local electromagnetic field because of localized surface plasmon resonance (LSPR), making them suitable for dark-field microscope (DFM) detection [34,35,36]. Compared with the bright-field microscopy, DFM has the innate advantages of low background, non-photobleaching, which guarantees the high signal-to-noise ratio [37, 38]. Therefore, many DFM assays employed AuNPs as single-entity microreactors for biomarker analysis by controlling their size, shape, and chemical modification. For example, Huang’s group realized the highly sensitive detection of miR21 by the reduced signal background through vertical polarization excited DFM (Fig. 4d) [36]. The LSPR property of gold nanorods (AuNRs) displayed a pronounced green color with low scattering intensity, which was adjusted to red color with strong scattering intensity when the core-satellite AuNP assembly structure was constructed on the side of AuNRs through a catalyzed hairpin assembly (CHA) circuit in the presence of miR21. The proposed strategy could not only realize the sensitive detection of miR21 with a limit of detection (LOD) as low as 2 pM but also achieve the high spatial resolution imaging of cancer cells.
It should be mentioned that the nanoparticles are more difficult to be separated compared with the microparticles. And because of the relatively smaller size, the optical signal of nanoparticle-based single-entity assays is relatively weak, which always need the introduction of special detecting instruments, such as TIRFM and DFM. In addition, it is notable that researches also reported single entities, including QDs and AuNPs, can be directly adopted as the signaling probes for the analysis of biomarkers in target-induced probe release manners [39,40,41,42]. However, these strategies are beyond the scope of this review, in which only the single entities employed as the reaction carries are included.
Generally, in the analog optical counting-based single-entity bioassays, the targets are evenly allocated on the single-entity microreactors, which allows for the high-sensitive detection of target molecules by analyzing the overall optical signals on each entity. The mean optical signal intensity of all the interrogated entity population is statistically analyzed to quantify the target level. As such, the heterogeneities among the individual target molecules cannot be differentiated. And the ultra-low content of target may not be accurately detected because the signal induced by trace amount of target molecules is always weak, which may not be discernable from the inevitable non-specific background signals. Fortunately, the optical single-entity strategy has also permitted digital sensing formats, which is a great leap forward the new-generation bioassays.
Single-entity counting-based digital bioassays
Digital bioassays represent a new class of bioanalytical methods that enable the highly sensitive detection of biomolecules [4,5,6,7]. Different from analog bioassays, in digital assays, the reaction solution containing the extremely low concentrations of target molecules is compartmentalized into a lot of entities acting as microreactors, which is in a one/none target-to-one microreactor manner [43,44,45]. According to Poisson distribution, when the number of target molecules is far less than that of the microreactors, most of the microreactors contain up to one single target. Specifically, the microreactors containing one target show positive signal and recorded as “1,” while the ones without target are recorded as “0.” Therefore, the number/concentration of the target can be determined by counting the entities that show positive or negative signals.
Superior to the conventional analog assays, digital assays have plenty of inherent advantages. Firstly, in the digital assays, each target molecule is confined in a small volume of the entity to initiate an individual reaction, causing the enrichment of signal in a fL~nL volume that could prominently improve the detection sensitivity. Therefore, digital assays have great potential in detecting ultra-low concentration of biomarkers [6, 7, 43]. Secondly, the digital assays can reach a wide dynamic range of over 3~4 orders of magnitude by preparing thousands or even millions of microreactors. It is because that digital assays solely depend on counting the number of single target molecule-induced positive entities for signal readout, and the quantification result is not subject to the fluctuation of signal intensity in each single microreactor [4, 5]. Last but not least, because the biomarker targets are individually divided into the independent microreactors, digital assays allow the study of the heterogeneity of biomarkers, which can hardly be achieved by bulk measurement-based analog assays [6, 7]. In this section, digital bioassays are categorized as traditional digital bioassays that rely on the sealed microchambers and the recently developed microchamber-free digital bioassays.
Digital bioassays in sealed microchambers
The digital bioassay requires the target molecules in the reaction solution to be partitioned into equal small entities. After the reaction for signal conversion inside the individual entities, the signals in each of them can be interrogated. Up to now, most of the entities adopted for digital bioassays are microchambers. The emulsion droplets and microwell arrays are two dominant microchambers used in digital bioassays.
Emulsion droplets as single entities for digital bioassays
The water-in-oil emulsion droplets are commonly used as the single microchamber entities in digital bioassays, which can be formed with the help of a microfluid system or direct vortex of aqueous phase and oil phase mixture. In this review, the emulsion droplet digital bioassays are classified on the basis of the involvement of beads.
-
(1)
Water-in-oil emulsion droplet platforms
The droplet digital PCR (ddPCR) is one of the most prominent approaches that uses water-in-oil droplets as the microreactors to realize high-throughput digital PCR in a low-cost and practical fashion [46,47,48,49]. The target biomarker molecules and PCR reagents are partitioned into ∼20,000 or more monodisperse droplets, resulting in zero or one target molecule in each droplet. These droplets act as the independent container for PCR amplification and each droplet can be assigned as positive (1) or negative (0) according to a fluorescence intensity threshold. Finally, the copy number/concentration of the target molecules is determined by the number of positive and negative entities by Poisson statistics.
As the pioneer to invent ddPCR, Hindson’s group successfully realized the estimation of DNA copy number with high-throughput by using the early access beta-prototype ddPCR platform (Bio-Rad, Pleasanton, CA) in 2011 [46]. After then, to improve the ddPCR platform’s sensing performance, they evaluated key factors that could influence the reliability of results obtained from their ddPCR system and assessed the linearity of the response and the precision over a dynamic range of the 20,000-droplet assay [47]. Until now, the ddPCR platform has been successfully used in virous biomarker analysis, including DNA, RNA, SNP, virus, and exosomes. For example, Tian et al. reported a highly sensitive and precisely ddPCR method for miRNA detection (Fig. 5a) [50]. In the presence of miRNAs, two target-specific ssDNA probe A and probe B were ligated with each other under the catalysis of T4 RNA ligase 2. Then, the ligation products were partitioned into the droplets to conduct PCR, resulting in the cleavage of TaqMan probes and enrichment of fluorescence signal. Lastly, the amounts of target miRNA were reflected by the number of fluorescence-positive droplets. Yang’s group developed a dual-aptamer activated proximity-induced ddPCR strategy for the quantitation of tumor-derived exosomal PD-L1 [51]. As shown in Fig. 5b, when introducing the target exosomes that express EpCAM and PD-L1, the two aptamers would simultaneously bind on the surface of exosomes and get close to each other. With the help of a connector probe and the ligase, the extended ends of the two aptamers were closely ligated via proximity ligation assay. After ligation, ddPCR was performed to absolutely quantify the ligation products, which, in turn, reflected the number of tumor derived exosomal PD-L1. Considering the ravages of the COVID-19 pandemic, Bu et al. furtherly demonstrated a novel ddPCR assay that was successfully applied to the detection of low copy number SARS-CoV-2 [52]. In this assay, low-cost droplet printing chips were introduced to generate a lot of well-dispersed and aligned PCR substrate droplets on a normal glass substrate without surface modification. After covering with a layer of oil, PCR thermal cycling and imaging were directly performed by using flat-panel PCR machines and microscopes that are routinely available in bio and medical labs. This system generated high-quality digital PCR data for SARS-CoV-2 analysis and broadened the application of the ddPCR platform.
Emulsion droplet digital platforms. a Illustration of the ddPCR-based miRNA assay. Reprinted with permission of [50]. Copyright 2016, American Chemical Society. b Working principle of the highly sensitive quantification of tumor-derived exosomal PD-L1 using proximity-induced ddPCR strategy. Reprinted with permission of [51]. Copyright 2021, Wiley-VCH Verlag GmbH & Co. KGaA
To overcome the technical barriers in developing ddPCR systems, a lot of methods have been proposed for their optimization. Firstly, the traditional droplet generation process has to rely on microfluid devices, which is a barrier that impedes the adoption of ddPCR, particularly in low resource setting-laboratories or ordinary biolabs. To solve this issue, Hatori et al. reported a new droplet-generated method by vortexing the mixture of water phase and oil phase [53, 54]. For example, they reported an approach for the accurate quantification of SARS-CoV-2 sequences by vortexing the target DNA and PCR reagents to prepare droplets (Fig. 6a) [54]. This method allows fast and scalable digital PCR without microfluidics, which generates the droplets by vortexing and obtains the results by common bulk techniques (electrophoresis and qPCR). Secondly, in the droplet generation procedure, surfactant is essential to stabilize the droplets. It is because that the water-in-oil droplets containing the targets and various biological agents must endure rapid temperature cycling for PCR. However, the surfactant can probably induce the crosstalk of positive and negative droplets, which would ultimately result in inaccurate detection [55, 56]. To address this issue, Yang’s group proposed the use of colloidosomes self-assembled from fluorinated silica nanoparticle (F-SiO2 NPs) [57]. As illustrated in Fig. 6b, the F-SiO2 NPs could self-assemble at the water-in-oil droplet interface when they were mixed with the emulsion droplet, forming extremely stable colloidosomes. After PCR amplification inside the colloidosomes, the number of bright and dark (positive and negative) colloidosomes was counted to evaluate the concentration of nucleic acid biomarker. Thirdly, when the reagent is altered, the emulsion droplets tend to be fragile, which inhabit the versatility of the emulsion droplet-based assays. Considering this, many groups developed more robust systems for digital analysis. For example, Zhu et al. reported a novel PEG hydrogel bead-based platform, in which uniform nanoliter-sized hydrogel beads (Gelbeads) were generated as microreactors [58]. As shown in Fig. 6c, the Gelbeads were developed by using a convenient and disposable device made of needles holding aqueous reaction mixture and the microcentrifuge tubes holding the oil phase. With centrifugal acceleration, the aqueous phase was forced into the fluorinated oil phase to form numbers of Gelbeads. By adding PCR and LAMP reagents in the aqueous phase, this thermally stable Gelbead-based system was successfully used in the single cell phenotyping and biomarker detection. This design allows for easy adoption of stable microreactor entities without expensive and complicated equipment. Finally, the most widely used amplification method in the emulsion-based digital assay is PCR, which requires complicated manipulations such as thermal cycling. And for the detection of RNA, extra reverse transcription procedures or other complexed design should be employed before PCR. Therefore, other efficient nucleic acid amplification strategies were introduced into the emulsion droplet to achieve the simplified operation [59,60,61]. For example, Zhou’s group employed clustered regularly interspaced short palindromic repeats (CRISPR)/Cas 13a system to droplet-based digital assay for the single-molecule RNA diagnostics (Fig. 6d) [59]. They partitioned the mixture of target RNA, Cas13a, crRNAs, and fluorescently labeled RNA reporters into thousands of monodisperse droplets. According to their design, the target miRNA could bind with the crRNA to activate the trans cleavage ability of Cas13a. In this way, the fluorescent molecules were liberated after cleaving the self-quenched RNA reporter, yielding high fluorescent signal confined in ultrasmall volume droplets. At last, RNA quantification was realized by digitally counting the fluorescent-positive droplets via fluorescence imaging.
-
(2)
Bead-involved emulsion droplet platforms
Extension of the emulsion droplet platforms. a Principal illustration of the emulsion droplet generation by vortexing the mixture of water phase and oil phase. Reprinted with permission of [54]. Copyright 2021, American Chemical Society. b Principle of the colloidosome-based digital PCR system. Reprinted with permission of [57]. Copyright 2019, American Chemical Society. c Schematic of PEG hydrogel bead-based platform. Reprinted with permission of [58]. Copyright 2021, American Chemical Society. d Workflow of the CRISPR/Cas 13a system-based droplet digital assay. Reprinted with permission of [59]. Copyright 2020, American Chemical Society
Despite the efforts devoted to this area, the background signal derived from homogeneous reactions or the complex biological samples can hardly be completely eliminated, which may become a main obstacle to the emulsion droplet platforms. Considering this, many groups introduced the beads into the emulsion droplet systems to serve as the medium to capture the biomarkers and separate them from the background media/signal [62,63,64].
BEAMing is one of the most famous bead-involved emulsion droplet digital bioassay methods, which is named on the basis of four of its principal components (beads, emulsion, amplification, and magnetics) [65,66,67]. As shown in Fig. 7a, in the BEAMing system, the target nucleic acid, PCR primer-coated magnetic beads, and PCR reagents are co-encapsulated in the emulsion droplets [65]. After the PCR amplification on the beads, the beads are released by breaking the droplet and then labeled by fluorescent probe via nucleic acid hybridization. Eventually, FCM is used to count the fluorescence-positive beads. With this design, the BEAMing system was successfully used in the detection of genetic variations and the quantification of mutations in the plasma of patients. Besides the PCR amplification, other efficient nucleic acid amplification strategies were also introduced in the BEAMing system. Li et al. introduced RCA to the BEAMing system to increase the number of copies bound to the beads by more than 100-fold (Fig. 7b) [68]. This allowed discrimination of mutant and wild-type sequences even when they were present at ratios less than 1:10,000. In another work by Chen et al., they have developed a novel BEAMing platform by combining emulsion microreactors, single-molecule magnetic capture, and on-bead loop-mediated isothermal amplification (LAMP) [69]. As illustrated in Fig. 7c, in this work, the target DNA initiated the LAMP amplification on the beads. Finally, the target DNA was amounted by counting the positive beads with FCM.
BEAMing-related platforms. a Schematic of BEAMing. Reprinted with permission of [65]. Copyright 2003, The National Academy of Sciences. b Principle of the RCA-based BEAMing system for the quantification of rare sequence variants. Reprinted with permission of [68]. Copyright 2006, Springer Nature Customer Service Centre GmbH: Springer Nature. c Illustration of a BEAMing LAMP system for digital DNA detection. Reprinted with permission of [69]. Copyright 2018, Royal Society of Chemistry
The digital protein analysis generally relies on the immunoassay to bring an enzyme molecule on one single bead [70,71,72]. After distributing the beads into emulsion droplets, the enzyme molecules catalyze the fluorescein substrates to lighten the emulsion droplets. For example, Shim et al. described a multilayered microfluidic device to enable the generation and manipulation of highly monodispersed femtoliter droplets for the single-molecule-counting immunoassay and single enzyme analysis (Fig. 8a) [70]. In the presence of antigen target, the single sandwich immunoreaction occurred to introduce the biotinylated detection antibody on the individual beads, and then the single STV-conjugated β-galatosidase (β-Gal) could bind on the single beads. The femtoliter droplets produced by a microfluid device were used to encapsulate the single beads coated with β-Gal and fluorogenic substrate fluorescein. By counting the fluorescence-positive droplets, the single-molecule-counting immunoassay was achieved. Liu et al. developed an immunosorbent digital assay for the qualification of target exosomal proteins by using droplet microfluidics (Fig. 8b) [71]. The single exosomes were firstly distributed to beads in a one-to-one manner via sandwich immunoreaction, causing the β-Gal bound on bead. The beads loaded with target exosome were further isolated and encapsulated into a sufficient number of droplets to ensure only one single bead was encapsulated in a droplet. Finally, in the droplets containing the beads with exosome immunocomplex, the substrate was catalyzed by β-Gal to emit fluorescein. On the basis of the statistics of the fluorescent droplets, the target exosome concentration down to ~10−17 M could be calculated. The simultaneous detection of a large number of targets is also important in digital bioassays. Considering this, Yelleswarapu et al. proposed a multiplexed, digital protein droplet detection by using fluorescence-coded beads (Fig. 8c) [72]. In this assay, each kind of fluorophore encoded beads were labeled with certain kinds of capture antibodies. After the specific immunoreaction, the β-Gal enzymes were immobilized on the beads. Finally, the individual beads were encapsulated into droplets and read out if they had captured a single target protein.
Bead-involved emulsion droplet platforms for protein analysis. a Principle of single enzyme molecule analysis with monodispersed femtoliter droplets. Reprinted with permission of [70]. Copyright 2013, American Chemical Society. b Schematic showing the droplet digital qualification of target exosomal proteins by using droplet microfluidic devices. Reprinted with permission of [71]. Copyright 2018, American Chemical Society. c Illustration of multiplexed, digital protein droplet detection by using fluorescence-coded beads. Reprinted with permission of [72]. Copyright 2019, The National Academy of Sciences
Microwell array-based digital bioassays
Etching and microfabrication techniques allow for the design of uniform and precisely positioned microwell arrays at femtoliter level. The microwell arrays enable large-scale experiments because they can be reused by rinsing and refilling [73, 74]. Therefore, microwell technology brings a new breakthrough to digital bioassays.
Walt’s group pioneered optical-fiber bundles to create high-density microwell arrays [75]. Optical-fiber bundles consisted of a few thousands to 100,000 individual fiber-optic cores with diameters between 2 and 20 μm. The core material was selectively etched by acid to create an array of uniform fL microwells with a density of approximately 25,000 mm−2 on one end of the optical-fiber bundle for loading fluorescent probes. Meanwhile, the other end of the fiber bundle was connected to an epifluorescence microscope. After launching the excitation light into the entire array, the returning emitted light was filtered to remove any scattered excitation light and then detected by a CCD camera. They employed the arrays of femtoliter-sized reaction microwells for the detection of proteins, which termed as single-molecules arrays (Simoa) [76]. As shown in Fig. 9a, in the first step of Simoa, a sandwich immunocomplex was formed on a single MB, and the immunocomplex was labeled with a β-Gal enzyme. Then, the MBs were loaded into the array of femtoliter-sized wells. After sealing the loaded arrays against a rubber gasket in the presence of fluorogenic enzyme substrate, each MB was isolated in a femtoliter-volume reaction microwell. Beads possessing a single enzyme-labeled immunocomplex generated a high concentration of fluorescent product in the 50-fl microwells. In contrast, the microwells without loading MBs showed fluorescence-negative. By distinguishing the number of fluorescence-positive microwells, subfemtomolar protein targets could be digitally detected. With this system, various types of biomarkers, including miRNAs and virus, were successfully analyzed [77,78,79,80,81,82]. Although the Simoa-based strategies can achieve efficient analysis of biomarkers, they subject to the low sample loading and the requirement of special instruments, which hamper their widespread application.
Microwell array-based digital bioassays. a Principle of the Simoa workflow. Reprinted with permission of [76]. Copyright 2010, Springer Nature Customer Service Centre GmbH: Springer Nature. b Illustration of PDMS/PAA coverslip microreactor system. Reprinted with permission of [84]. Copyright 2005, Springer Nature Customer Service Centre GmbH: Springer Nature
Besides the microwell arrays used in Simoa, Noji’s group develop another kind of microwell arrays by sandwiching the sample solution between a poly-dimethylsiloxane (PDMS) or polyacrylamide (PAA) gel with many microcavities on the surface and a coverglass (Fig. 9b) [83,84,85]. For example, in 2005, they developed a silicone device presenting a large array of micrometer-sized cavities [84]. They used the PDMS that was transparent to visible light and spontaneously adhesive to smooth surfaces to build up this device. Specifically, a cylindrical silicon wafer array was prepared and used as the master template to produce series of identical PDMS sheets by molding. Then, the sample solution was dropped in between of a microscope glass slide and the patterned PDMS sheet, forming large number of femtoliter microwells. By using β-Gal and horseradish peroxidase (HRP) as model enzymes, such microwells allowed the quantitative detection of the products yielded by a single enzyme molecule. In addition to PDMS, the PAA gel can also be employed for fabricating microwells, which were used in the restriction enzyme assay [85]. Nevertheless, the complexed steps for the preparation of microwell arrays may hamper their prevalence.
Droplet array-based digital bioassays
Besides the methods employed emulsion droplets or microwells as microchambers, many droplet array-based digital technologies have also been developed by combining the advantages of emulsion droplets and microwells [86,87,88,89,90,91].
Typically, Noji’s group has developed an emerging femtoliter droplet array (Fig. 10a) [86]. Firstly, the hydrophobic polymer of carbon-fluorine (CYTOP) was spin-coated on a cleaned coverglass. Then the resist-patterned substrate surface was dry-etched with O2 plasma by a reactive ion etching system to expose the hydrophilic SiO2 glass surface. After covered with aqueous solution, oil was flowed into the substrate surface. Because the oil employed here had a higher density than water, the hydrophilic SiO2 glass surface retained the aqueous solution, while the hydrophobic surface was covered with oil. As a result, many droplets were formed simultaneously. With this droplet array, they successfully measured the single-molecule antigen (Fig. 10b) [87]. In this assay, the MBs coated with capture antibody were introduced. After the antigen-specific immunoassay, the β-Gal was immobilized on the MBs. And then, the β-Gal-enriched MBs were injected into the digital counting device and trapped into the droplets in an array to catalyze the chromogenic reaction of fluorescent substrate. The antigens were digitally quantified by monitoring the fraction of bright droplets. Furthermore, they assembled a flow cell with the droplet-array system to build up a multiconditional and multiparametric digital bioassay platform, which was capable of exchanging solution between the microreactors [90]. As shown in Fig. 10c, in another work, they immobilized single analytes in arrayed femtoliter reactors and sealed them with airflow. By replacing the solution with buffer or the next sample, and repeating the sealing procedure, perfect solution exchanges were successfully performed. With this new multidimensional digital assay, the quantitative determination of inhibitor sensitivities of single influenza A virus particles and single ALP was achieved.
Droplet array-based digital bioassays. a Schematic drawing of droplet array preparation procedure developed by Noji’s group. Reprinted with permission of [86]. Copyright 2010, Royal Society of Chemistry. b Schematic illustrations of the concept of digital analysis of single-molecule antigen based on droplet array. Reprinted with permission of [87]. Copyright 2012, Royal Society of Chemistry. c Flow cell integrated droplet array device for the multiconditional and multiparametric analysis of biomarkers. Reprinted with permission of [90]. Copyright 2021, American Chemical Society. d Single molecule protein detection using droplet array-based digital ELISA strategy. Reprinted with permission of [91]. Copyright 2020, American Chemical Society
In addition, in order to improve the bead loading efficiency of Simoa, Walt’s group also developed a droplet array-based digital ELISA strategy for single protein molecule detection (Fig. 10d) [91]. Following the principle of Poisson distribution, at most one β-Gal enzyme was immobilized on a single MB. Then, the MBs were isolated into picoliter-sized droplets such that each droplet could fit one bead at most. The droplets were then loaded into a chamber at a monolayer to form droplet arrays. Eventually, an imaging system was adopted to identify the fluorescence-positive MBs which could reflect the number of protein target.
Although the droplet array-based digital arrays combine the advantages of emulsion droplet and microwell, they also have the disadvantages such as the specialized droplet preparation and complexed microwell fabrication.
Microchamber-free digital bioassays
Although the microchamber-based digital assays have been widely used in the ultrasensitive analysis of biomarkers, they may suffer from some drawbacks. For example, with regard to the emulsion droplet-based digital bioassays, the emulsion droplet is fragile. And specialized devices and accessories are needed for preparing the droplets, causing the large size and complicated measurement of the systems. As for Simoa, its sensitivity is limited by low sampling efficiencies. According to literature reports, only about 5% of the total number of beads can be loaded into the microwells by gravity and ultimately be analyzed [92]. And for most of the microchamber-based digital assays, complicated microchamber preparation procedure is required to make sure each microchamber is completely sealed in order to prevent the cross-reaction between the adjacent chambers. Given this, a series of microchamber-free digital assays were developed for the analysis of biomarkers [92,93,94,95,96,97,98,99]. Among them, the FCM-based ones have outstanding advantages to allow for digital detection of biomolecules because FCM could rapidly obtain the fluorescence signals on the single entities and decode the entities’ size information with high throughput. It is notable that the FCM-based digital assays have essential differences from the FCM-based analog ones mentioned above. On the one hand, in the FCM-based digital assays, the number of target molecules is normally far less than that of the microreactors and at most one target molecule is captured on/in each microreactor according to Poisson distribution. In contrast, in a typical FCM-based analog assay, the target molecules are averagely distributed to the microreactors because the number of them is far more than that of the microreactors. On the other hand, considering the mode of signal readout, in the FCM-based analog assays, the mean fluorescence intensity of a number of microreactors is obtained and analyzed to reflect the concentration of target molecules, whereas in the digital ones, the absolute fluorescence intensity of the microreactors is not essential because the concentration of the target molecules is determined by counting the number of positive microreactors according to a pre-set threshold.
Akama et al. presented a novel digital ELISA that did not require sealed microchambers [94]. As shown in Fig. 11a, by adjusting the concentration of MB, at most one HRP enzyme was immobilized on a single MB after a sandwiched immunoreaction. Then, the biotin-labeled tyramide substrate reacted with HRP to convert into biotin-labeled tyramide radical, which could bind to the aromatic moieties of the protein on the MBs. Then the STV-labeled fluorescent dyes bound to biotin deposited on the MBs. In this way, as low as 0.09 mIU/mL (139 aM) antigen target with a wide dynamic range was digitally analyzed by counting the number of fluorescent MBs with FCM. Although this method achieved the microchamber-free digital analysis of the target biomarker, the signal strength of some positive beads may be difficult to be completely distinguished from the negative ones because the weak fluorescent signals deposited on a very small area of each MB, leading to an inaccuracy of positive rate.
Microchamber-free digital bioassays. a Schematic illustration of a typical droplet-free digital ELISA system. Reprinted with permission of [94]. Copyright 2016, American Chemical Society. b Schematic illustration of the workflow of the digital FCM platform for miRNA detection. Reprinted with permission of [95]. Copyright 2020, Royal Society of Chemistry. c Schematic illustration of the RCA-assisted FCM for profiling exosomal surface proteins. Reprinted with permission of [96]. Copyright 2021, American Chemical Society. d Schematic of dropcast Simoa strategy for the ultrasensitive detection of protein. Reprinted with permission of [92]. Copyright 2020, American Chemical Society. e Schematic work principle of the single MNP-confined, click chemistry-actuated ddWalker for digital miRNA analysis. Reprinted with permission of [98]. Copyright 2020, American Chemical Society. f Design principle of the digital T4 PNKP assay. Reprinted with permission of [99]. Copyright 2021, American Chemical Society
Considering this, researchers have integrated various nucleic acid amplification methods with the microchamber-free digital bioassays to enrich more optical signals in the presence of only one target molecule. For example, as shown in Fig. 11b, our group developed a digital FCM platform toward the precise quantification of target miRNAs [95]. In this platform, at most one target-initiated ligation product with 3′-OH termini was bound on one single MB via the click nucleic acid ligation (CNAL) reaction. After then, the single ligation product induced the TdT extension and HCR amplification to enrich fluorescent molecules on single MBs. Eventually, as low as attomolar level of target miRNA was detected by counting the fluorescence-positive MBs with FCM. Similarly, Li’s group proposed the digital FCM system for the profiling of exosomal surface proteins by introducing RCA reaction (Fig. 11c) [96]. In this design, the specific anti-CD63 antibody-conjugated magnetic MBs were utilized to capture exosomes. Then the ssDNA that comprises an exosomal surface protein-specific recognition aptamer and an RCA trigger was introduced to bind with the target exosome. After the RCA reaction, plenty of fluorescent probes were hybridized with the RCA products to amplify the fluorescent signals. The count of fluorescence-positive MBs was proportional to the amount of exosomal membrane proteins, allowing quantitative analysis by FCM. Besides the digital FCM system, Walt’s group developed an RCA-based dropcast Simoa strategy, called dSimoa, for the ultrasensitive detection of attomolar protein concentrations (Fig. 11d) [92]. In this approach, single immunocomplex sandwich structures formed on the antibody-coated MBs. Then, a STV-DNA conjugate containing an RCA primer was bound on the MBs via the interaction between biotinylated detector antibody and STV-DNA. After RCA reaction, the product DNA concatemer on the MBs hybridized with fluorescently labeled DNA probes for visualization. Finally, the MBs were simply dropcast onto a microscope slide and dried into a monolayer film for digital signal readout. The dSimoa platform achieved attomolar LOD, with an up to 25-fold improvement in sensitivity over Simoa.
Compared with the normally used microbeads, for smaller entities, less optical molecules are required to lighten them up for discriminating them from the negative ones. Considering this, we devised a versatile single MNP-confined, click chemistry-actuated digital DNA walker (ddWalker) machine for the analysis of miRNA (Fig. 11e) [98]. By controlling the number of MNPs, at most one single DNA walking leg was immobilized on a single MNP via the miRNA-mediated CNAL reaction. In the presence of nicking enzyme, the fluorescence-quenched molecular beacon tracks fixed on the MNPs was cleaved to release the quencher and the metastable walking legs. Then, the walking legs hybridized with another molecular beacon tracks to initiate the next cycle of nicking cleavage, resulting in fluorescent molecules enriched on the MNPs. Finally, by digitally counting the number of fluorescence-on MNPs with TIRFM, femtomolar-level quantification of miRNA was realized.
In addition, we implemented the analysis of enzyme activity by using digital FCM system [99]. As shown in Fig. 11f, we proposed a FCM method for the digital quantification of T4 polynucleotide kinase phosphatase (T4 PNKP) based on the extraordinary space-confined enzymatic feature of T4 PNKP that each T4 PNKP-catalyzed reaction could be spatially self-confined on a single MB. In this design, T4 PNKP dephosphorylated the 3′-PO4 DNA substrates step by step on a single MB until the substrates on the surface of the residential MB were exhausted. In this way, the 3′-PO4 groups of the DNA substrates were converted into the 3′-OH groups, which could be recognized by TdT to initiate the DNA polymerization and the hybridization of fluorescent probes. The proportion of fluorescence-positive MBs was investigated with FCM to digitally monitor the single enzyme activity. As a result, as low as 1.28 × 10−10 U/μL T4 PNKP was clearly detected with this strategy. This study also suggested that by using T4 PNKP as the signaling enzyme probe, the novel digital mechanism is promising to be extended to digital ELISA or other microchamber-free digital bioassays.
Microchamber-free digital strategies have a good applicability. Therefore, we prospect that the development of microchamber-free digital bioassays will further promote the analysis of biomarkers.
Perspective
Although researchers have devoted considerable efforts to the single-entity optical bioanalysis, further works are still required in regard to their applications in fundamental biological studies and clinical diagnosis. Considering the inherent advantages of digital assays compared with the analog ones, especially the ultra-high sensitivity and the ability to discriminate the intermolecular heterogeneity, the future trend of the single entity-based digital assay is prospected from the following aspects.
Compared with the microchamber-based digital assays, the microchamber-free ones have shown great potential in the real-word applications especially in routine biolabs, because they avoid the usage of fragile droplets, complex microchamber fabrication procedures, and specialized instruments. Therefore, it is vital to exploit simpler and more efficient microchamber-free systems. As mentioned above, FCM is an easily accessible instrument widely equipped in hospitals and routine labs. Although many research groups have employed conventional FCM for the MB-based microchamber-free digital analysis of biomarkers, the FCM-based digital assay is still in its infancy. In order to improve the digital analysis performance of single entity-based digital assays, it is still highly desired to develop emerging target-responsive spatial-confined reaction mechanisms and robust amplification methods on the MB surface for higher sensitivity and simple and automatic instruments for the convenience of signal readout.
The next-generation automatic microfluid system, which can integrate target capture, signal generation, and signal readout, should be the focus of future work. At present, for the convenience of operation, the entity-based microfluid technologies generally employ entities of large sizes (e.g., a few to hundreds micrometer of droplet or microwell in commercialized systems). It is generally acknowledged that single entity with a smaller diameter will promote the detection performance because they require fewer fluorescent molecules on the surface to make them bright enough to be discernable to the negative ones. Therefore, the microfluid devices that can manipulate and easily count small size of entities may help to achieve the single molecule level detection with quite simple signal amplification or even without any signal amplification strategies.
Moreover, we suppose that besides the ones mentioned above, more types of ultrasensitive optical-based signaling technologies, such as SERS and chemiluminescence, can be introduced to the single-entity bioassays. For example, Li et al. developed a digital nanopillar SERS platform for the digital detection of single cytokine [100]. In this design, they fabricated gold nanopillar as microreactor, which acted as the small compartment to capture and confine the individual cytokine. After the specific immunoreaction, the SERS nanotags, which were produced by co-conjugating Raman reporter and target antibody onto gold-silver alloy nanoboxes, were immobilized on the gold nanopillars. In this way, the cytokine target can be digitally measured by mapping the SERS signals. The digital nanopillar SERS assay achieved high sensitivity with down to attomolar-level LOD and multiplexed cytokine detection capability. However, this work only reported a preliminary design of a digital SERS platform. Compared to the mature digital platforms such as ddPCR and Simoa, the precision in biomarker digital analysis is not so satisfactory. Considering the inherent advantages of SERS signals that superior to the fluorescence signals, including single molecule sensitivity, high throughput, and high photostability, we predict that the digital SERS platforms can be promising supplements to the digital fluorescence systems after being combined with appropriate experiment design and reasonable algorithms. Furthermore, it is believed that the integration of other types of optical signal with single-entity assay will permit new breakthroughs in the field of biomarker analysis.
Besides designing new principles, employing emerging devices, and exploring more sensitive optical signaling technologies, the experimental details are also important factors to the accuracy and stability of the single entity-based bioassays. For one thing, the heterogeneities among different micro/nano-reactors may lead to varying signals as well as imprecise results. Fortunately, digital bioassays can reduce such influences to a large extent, because the biomolecule targets are quantified by counting the number of positive single entities according to a rationally set threshold, in which the variation of optical signal intensity on each single entity can be reasonably tolerated. Nevertheless, the negative impact of the heterogeneities among single entities cannot be thoroughly eliminated. Therefore, developing uniform, stable, and well-monodispersed micro/nano-reactors is still the key to the accurate application of single entity-based bioassays. For another, considering the variety of biomolecule targets, different labeling methods should be introduced into the single entity-based assays. Up to now, lots of labeling methods, including electrostatic adsorption, chemical crosslinking, biological orthogonal reaction, and biological affinity reaction, have been widely applied in biological recognition. In this regard, we believe that developing more robust and simple labeling methods is critical to guarantee the accuracy and stability of the single entity-based bioassays.
In addition to the technique aspects, we suggest more attention should be paid to the application of single entity-based digital assays. On the one hand, because the occurrence of disease is often accompanied with the change of more than one biomarker, it would be of great significance if new strategies can be developed to achieve the multiplexed analysis of biomarkers by using the conventional instruments or devices such as FCM. On the other hand, the heterogeneity phenomenon, such as the presence of rare mutations in a DNA sample [101], differences in catalytic activities of individual enzymes [102], and different telomere length in various stage of age-associated diseases [103, 104], widely exists in complex biological systems. Therefore, revealing the accurate pattern of the biomolecule heterogeneity in organisms or life processes is of great importance to precisely understanding of the occurrence and development of life process and disease, which cannot be achieved via the analog assays. Fortunately, digital assay has great potential in detecting the heterogeneity of the target molecules because of its ability to provide information at a single-molecule level. For example, Noji’s group established a digital droplet array-based single-molecule enzymatic assay [105]. They counted the number of enzyme molecules by profiling their single enzyme activity characteristics toward multiple substrates. They introduced the coumarin fluorogenic substrate, which had much shorter excitation and emission wavelengths than conventional substrate of fluorescein and resorufin. With this substrate, not only single enzyme activity was detected, but also multiplexed enzyme analysis was achieved. And if the biomarker molecule is telomere that is a highly relevant to age-associated diseases and cancer, its length is in a large heterogeneity among chromosomes. As is reported, Luo et al. developed the single telomere absolute-length rapid assay, a novel high-throughput digital real-time PCR approach for rapidly measuring the absolute lengths and quantities of individual telomere molecules [106]. This technique provided the accuracy and sensitivity to uncover associations between telomere length distribution and telomere maintenance mechanisms in cancer cell lines and primary tumors. As mentioned above, digital assay is an important tool for detecting the heterogeneity of the target molecules. Therefore, developing more advanced single entity-based digital bioassays for monitoring the heterogeneity between target molecules is another direction toward the clinical diagnosis in the future.
Conclusions
Over the past several years, single-entity optical counting bioassays have experienced very rapid growth. Superior to the conventional ensemble measurements, single-entity optical approaches enable the examination of individual entities within an entire population at fM or even aM range. In this review, the design principles and the analytical performance of the optical counting bioassays employing single entities as reactors have been summarized. We have also pointed out the further trends in this area to promote the analytical performance of the single-entity counting-based assays toward clinical diagnosis and therapy. It is believed that with the consistent efforts devoted to this area, the practical application of the single-entity counting assays will meet its prosperity in the near future.
References
Ven K, Vanspauwen B, Pérez-Ruiz E, Leirs K, Decrop D, Gerstmans H, Spasic D, Lammertyn J. Target confinement in small reaction volumes using microfluidic technologies: a smart approach for single-entity detection and analysis. ACS Sens. 2018;3(2):264–84. https://doi.org/10.1021/acssensors.7b00873.
Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc Natl Acad Sci USA. 2004;101(2):534–9. https://doi.org/10.1073/pnas.2636333100.
Ziemba BP, Li J, Landgraf KE, Knight JD, Voth GA, Falke JJ. Single-molecule studies reveal a hidden key step in the activation mechanism of membrane-bound protein kinase C-α. Biochemistry. 2014;53(10):1697–713. https://doi.org/10.1021/bi4016082.
Walt DR. Optical methods for single molecule detection and analysis. Anal Chem. 2013;85(3):1258–63. https://doi.org/10.1021/ac3027178.
Zhang Y, Noji H. Digital bioassays: theory, applications, and perspectives. Anal Chem. 2017;89(1):92–101. https://doi.org/10.1021/acs.analchem.6b04290.
Farka Z, Mickert MJ, Pastucha M, Mikušová Z, Skládal P, Gorris HH. Advances in optical single-molecule detection: en route to supersensitive bioaffinity assays. Angew Chem Int Ed. 2020;59(27):10746–73. https://doi.org/10.1002/anie.201913924.
Gooding JJ, Gaus K. Single-molecule sensors: challenges and opportunities for quantitative analysis. Angew Chem Int Ed. 2016;55(38):11354–66. https://doi.org/10.1002/anie.201600495.
Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol. 2017;37(2):163–76. https://doi.org/10.3109/07388551.2015.1128876.
Wilkerson MJ. Principles and applications of flow cytometry and cell sorting in companion animal medicine. Vet Clin North Am Small Anim Pract. 2012;42(1):53–71. https://doi.org/10.1016/j.cvsm.2011.09.012.
Fraga M, Vilarino N, Louzao MC, Rodriguez P, Campbell K, Elliott CT, Botana LM. Multidetection of paralytic, diarrheic, and amnesic shellfish toxins by an inhibition immunoassay using a microsphere-flow cytometry system. Anal Chem. 2013;85(16):7794–802. https://doi.org/10.1021/ac401146m.
Li D, Wang Y, Lau C, Lu J. xMAP array microspheres based stem-loop structured probes as conformational switches for multiplexing detection of miRNAs. Anal Chem. 2014;86(20):10148–56. https://doi.org/10.1021/ac501989b.
Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363(1-2):71–82. https://doi.org/10.1016/j.cccn.2005.06.023.
Qiu L, Zhang Y, Liu C, Li Z. A versatile size-coded flow cytometric bead assay for simultaneous detection of multiple microRNAs coupled with a two-step cascading signal amplification. Chem Commun. 2017;53(20):2926–9. https://doi.org/10.1039/C7CC00618G.
Ren W, Liu H, Yang W, Fan Y, Yang L, Wang Y, Liu C, Li Z. A cytometric bead assay for sensitive DNA detection based on enzyme-free signal amplification of hybridization chain reaction. Biosens Bioelectron. 2013;49:380–6. https://doi.org/10.1016/j.bios.2013.05.055.
Zhang Y, Liu C, Sun S, Tang Y, Li Z. Phosphorylation-induced hybridization chain reaction on beads: an ultrasensitive flow cytometric assay for the detection of T4 polynucleotide kinase activity. Chem Commun. 2015;51(27):5832–5. https://doi.org/10.1039/C5CC00572H.
Qi Y, Qiu L, Fan W, Liu C, Li Z. An enzyme-free flow cytometric bead assay for the sensitive detection of microRNAs based on click nucleic acid ligation-mediated signal amplification. Analyst. 2017;142(16):2967–73. https://doi.org/10.1039/C7AN00989E.
Fan W, Qi Y, Qiu L, He P, Liu C, Li Z. Click chemical ligation-initiated on-bead DNA polymerization for the sensitive flow cytometric detection of 3'-terminal 2'-O-methylated plant microRNA. Anal Chem. 2018;90(8):5390–7. https://doi.org/10.1021/acs.analchem.8b00589.
Zhu L, Chen D, Lu X, Qi Y, He P, Liu C, Li Z. An ultrasensitive flow cytometric immunoassay based on bead surface-initiated template-free DNA extension. Chem Sci. 2018;9(32):6605–13. https://doi.org/10.1039/C8SC02752H.
Smith LD, Liu Y, Zahid MU, Canady TD, Wang L, Kohli M, Cunningham BT, Smith AM. High-fidelity single molecule quantification in a flow cytometer using multiparametric optical analysis. ACS Nano. 2020;14(2):2324–35. https://doi.org/10.1021/acsnano.9b09498.
Zhang X, Liu C, Wang H, Wang H, Li Z. Rare earth ion mediated fluorescence accumulation on a single microbead: an ultrasensitive strategy for the detection of protein kinase activity at the single-cell level. Angew Chem Int Ed. 2015;54(50):15186–90. https://doi.org/10.1002/anie.201507580.
Zhang X, Liu C, Sun L, Duan X, Li Z. Lab on a single microbead: an ultrasensitive detection strategy enabling microRNA analysis at the single-molecule level. Chem Sci. 2015;6(11):6213–8. https://doi.org/10.1039/C5SC02641E.
Chen D, Zhang X, Zhu L, Liu C, Li Z. All on size-coded single bead set: a modular enrich-amplify-amplify strategy for attomolar level multi-immunoassay. Chem Sci. 2022;13(12):3501–6. https://doi.org/10.1039/D1SC07048G.
Lu X, Ren W, Hu C, Liu C, Li Z. Plasmon-enhanced surface-enhanced Raman scattering mapping concentrated on a single bead for ultrasensitive and multiplexed immunoassay. Anal Chem. 2020;92(18):12387–93. https://doi.org/10.1021/acs.analchem.0c02125.
Lu X, Hu C, Jia D, Fan W, Ren W, Liu C. Amplification-free and mix-and-read analysis of multiplexed microRNAs on a single plasmonic microbead. Nano Lett. 2021;21(15):6718–24. https://doi.org/10.1021/acs.nanolett.1c02473.
Wang W, Tao N. Detection, counting, and imaging of single nanoparticles. Anal Chem. 2014;86:(1):2-14. https://doi.org/10.1021/ac403890n.
Ma F, Li Y, Tang B, Zhang CY. Fluorescent biosensors based on single-molecule counting. Acc Chem Res. 2016;49(9):1722–30. https://doi.org/10.1021/acs.accounts.6b00237.
Li CC, Hu J, Luo X, Hu J, Zhang CY. Development of a single quantum dot-mediated FRET nanosensor for sensitive detection of single-nucleotide polymorphism in cancer cells. Anal Chem. 2021;93(43):14568–76. https://doi.org/10.1021/acs.analchem.1c03675.
Wang ZY, Li DL, Tian X, Zhang CY. A copper-free and enzyme-free click chemistry-mediated single quantum dot nanosensor for accurate detection of microRNAs in cancer cells and tissues. Chem Sci. 2021;12(31):10426–35. https://doi.org/10.1039/D1SC01865E.
He D, Ho SL, Chan HN, Wang H, Hai L, He X, Wang K, Li HW. Molecular-recognition-based DNA nanodevices for enhancing the direct visualization and quantification of single vesicles of tumor exosomes in plasma microsamples. Anal Chem. 2019;91(4):2768–75. https://doi.org/10.1021/acs.analchem.8b04509.
Chan HN, Ho SL, He D, Li HW. Direct and sensitive detection of circulating miRNA in human serum by ligase-mediated amplification. Talanta. 2020;206:120217. https://doi.org/10.1016/j.talanta.2019.120217.
Wong KW, Xu D, He D, Wong MS, Li HW. Direct immunomagnetic detection of low abundance cardiac biomarker by aptamer DNA nanocomplex. Sens. Actuators B Chem. 2019;291:200–6. https://doi.org/10.1016/j.snb.2019.04.035.
Qi Y, Lu X, Feng Q, Fan W, Liu C, Li Z. An enzyme-free microRNA assay based on fluorescence counting of click chemical ligation-illuminated magnetic nanoparticles with total internal reflection fluorescence microscopy. ACS Sens. 2018;3(12):2667–74. https://doi.org/10.1021/acssensors.8b01169.
Feng Q, Zhai Y, Ren W, Liu C. Target extension-activated DNA walker on nanoparticles for digital counting-based analysis of microRNA. Chin J Chem. 2021;39(6):1471–6. https://doi.org/10.1002/cjoc.202000692.
Qi F, Han Y, Ye Z, Liu H, Wei L, Xiao L. Color-coded single-particle pyrophosphate assay with dark-field optical microscopy. Anal Chem. 2018;90(18):11146–53. https://doi.org/10.1021/acs.analchem.8b03211.
Wang F, Li Y, Han Y, Ye Z, Wei L, Luo HB, Xiao L. Single-particle enzyme activity assay with spectral-resolved dark-field optical microscopy. Anal Chem. 2019;91(9):6329–39. https://doi.org/10.1021/acs.analchem.9b01300.
Liu J, Yan H, Zhang Q, Gao P, Li C, Liang G, Huang C, Wang J. High-resolution vertical polarization excited dark-field microscopic imaging of anisotropic gold nanorods for the sensitive detection and spatial imaging of intracellular microRNA-21. Anal Chem. 2020;92(19):13118–25. https://doi.org/10.1021/acs.analchem.0c02164.
McFarland AD, Van Duyne RP. Single silver nanoparticles as real-time optical sensors with zeptomole sensitivity. Nano Lett. 2003;3(8):1057–62. https://doi.org/10.1021/nl034372s.
Gao P, Lei G, Huang C. Dark-field microscopy: recent advances in accurate analysis and emerging applications. Anal Chem. 2021;93(11):4707–26. https://doi.org/10.1021/acs.analchem.0c04390.
Ma F, Zhao N, Liu M, Xu Q, Zhang C. Single-molecule biosensing of alkaline phosphatase in cells and serum based on dephosphorylation-triggered catalytic assembly and disassembly of the fluorescent DNA chain. Anal Chem. 2022;94(15):6004–10. https://doi.org/10.1021/acs.analchem.2c00603.
Li CC, Chen HY, Hu J, Zhang CY. Rolling circle amplification-driven encoding of different fluorescent molecules for simultaneous detection of multiple DNA repair enzymes at the single-molecule level. Chem Sci. 2020;11(22):5724–34. https://doi.org/10.1039/D0SC01652G.
Li T, Wu X, Tao G, Yin H, Zhang J, Liu F, Li N. A simple and non-amplification platform for femtomolar DNA and microRNA detection by combining automatic gold nanoparticle enumeration with target-induced strand-displacement. Biosens Bioelectron. 2018;105:137–42. https://doi.org/10.1016/j.bios.2018.01.034.
Pei X, Hong H, Liu S, Li N. Nucleic acids detection for mycobacterium tuberculosis based on gold nanoparticles counting and rolling-circle amplification. Biosensors. 2022;12(7):448. https://doi.org/10.3390/bios12070448.
Liu H, Lei Y. A critical review: recent advances in “digital” biomolecule detection with single copy sensitivity. Biosens Bioelectron. 2021;177:112901. https://doi.org/10.1016/j.bios.2020.112901.
Huang Q, Li N, Zhang H, Che C, Sun F, Xiong Y, Canady TD, Cunningham BT. Critical review: digital resolution biomolecular sensing for diagnostics and life science research. Lab Chip. 2020;20(16):2816–40. https://doi.org/10.1039/D0LC00506A.
Noji H, Minagawa Y, Ueno H. Enzyme-based digital bioassay technology-key strategies and future perspectives. Lab Chip. 2022;22(17):3092–109. https://doi.org/10.1039/D2LC00223J.
Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–10. https://doi.org/10.1021/ac202028g.
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–11. https://doi.org/10.1021/ac202578x.
Miotke L, Lau BT, Rumma RT, Ji HP. High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. Anal Chem. 2014;86(5):2618–24. https://doi.org/10.1021/ac403843j.
Decraene C, Silveira AB, Bidard FC, Vallee A, Michel M, Melaabi S, Vincent-Salomon A, Saliou A, Houy A, Milder M, Lantz O, Ychou M, Denis MG, Pierga JY, Stern MH, Proudhon C. Multiple hotspot mutations scanning by single droplet digital PCR. Clin Chem. 2018;64(2):317–28. https://doi.org/10.1373/clinchem.2017.272518.
Tian H, Sun Y, Liu C, Duan X, Tang W, Li Z. Precise quantitation of microRNA in a single cell with droplet digital PCR based on ligation reaction. Anal Chem. 2016;88(23):11384–9. https://doi.org/10.1021/acs.analchem.6b01225.
Lin B, Tian T, Lu Y, Liu D, Huang M, Zhu L, Zhu Z, Song Y, Yang C. Tracing tumor-derived exosomal PD-L1 by dual-aptamer activated proximity-induced droplet digital PCR. Angew Chem Int Ed. 2021;60(14):7582–6. https://doi.org/10.1002/anie.202015628.
Bu W, Li W, Li J, Ao T, Li Z, Wu B, Wu S, Kong W, Pan T, Ding Y, Tan W, Li B, Chen Y, Men Y. A low-cost, programmable, and multi-functional droplet printing system for low copy number SARS-CoV-2 digital PCR determination. Sens Actuators B Chem. 2021;348:130678. https://doi.org/10.1016/j.snb.2021.130678.
Hatori MN, Kim SC, Abate AR. Particle-templated emulsification for microfluidics-free digital biology. Anal Chem. 2018;90(16):9813–20. https://doi.org/10.1021/acs.analchem.8b01759.
Sun C, Liu L, Vasudevan HN, Chang KC, Abate AR. Accurate bulk quantitation of droplet digital polymerase chain reaction. Anal Chem. 2021;93(29):9974–9. https://doi.org/10.1021/acs.analchem.1c00877.
Skhiri Y, Gruner P, Semin B, Brosseau Q, Pekin D, Mazutis L, Goust V, Kleinschmidt F, El Harrak A, Hutchison JB, Mayot E, Bartolo J-F, Griffiths AD, Taly V, Baret J-C. Dynamics of molecular transport by surfactants in emulsions. Soft Matter. 2012;8(41):10618–27. https://doi.org/10.1039/C2SM25934F.
Yin K, Zeng X, Liang X, Wei H, Zeng H, Qi W, Ruan W, Song Y, Yang C, Zhu Z. Crosstalk-free colloidosomes for high throughput single-molecule protein analysis. Sci China Chem. 2020;63(10):1507–14. https://doi.org/10.1007/s11426-020-9818-9.
Yin K, Zeng X, Liu W, Xue Y, Li X, Wang W, Song Y, Zhu Z, Yang C. Stable colloidosomes formed by self-assembly of colloidal surfactant for highly robust digital PCR. Anal Chem. 2019;91(9):6003–11. https://doi.org/10.1021/acs.analchem.9b00470.
Zhu Y, Li J, Lin X, Huang X, Hoffmann MR. Single-cell phenotypic analysis and digital molecular detection linkable by a hydrogel bead-based platform. ACS Appl Bio Mater. 2021;4(3):2664–74. https://doi.org/10.1021/acsabm.0c01615.
Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo Z, Han Z, Wang Z, Zhou X. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano. 2021;15(1):1167–78. https://doi.org/10.1021/acsnano.0c08165.
Wu H, Cao X, Meng Y, Richards D, Wu J, Ye Z, deMello AJ. DropCRISPR: A LAMP-Cas12a based digital method for ultrasensitive detection of nucleic acid. Biosens Bioelectron. 2022;211:114377. https://doi.org/10.1016/j.bios.2022.114377.
Yue H, Shu B, Tian T, Xiong E, Huang M, Zhu D, Sun J, Liu Q, Wang S, Li Y, Zhou X. Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level. Nano Lett. 2021;21(11):4643–53. https://doi.org/10.1021/acs.nanolett.1c00715.
Mou L, Hong H, Xu X, Xia Y, Jiang X. Digital hybridization human papillomavirus assay with attomolar sensitivity without amplification. ACS Nano. 2021;15(8):13077–84. https://doi.org/10.1021/acsnano.1c02311.
Wu H, Ma X, Chu Y, Qi X, Zou B, Liu Y, Zhou G. Digital nucleic acid signal amplification platform for highly sensitive DNA mutation analysis. Anal Chem. 2022;94(9):3858–64. https://doi.org/10.1021/acs.analchem.1c04765.
Gu Z, Sun T, Guo Q, Wang Y, Ge Y, Gu H, Xu G, Xu H. Bead-based multiplexed droplet digital polymerase chain reaction in a single tube using universal sequences: an ultrasensitive, cross-reaction-free, and high-throughput strategy. ACS Sens. 2022;7(9):2759–66. https://doi.org/10.1021/acssensors.2c01415.
Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100(15):8817–22. https://doi.org/10.1073/pnas.1133470100.
Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3(7):551–9. https://doi.org/10.1038/nmeth898.
Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–73. https://doi.org/10.1073/pnas.0507904102.
Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat Methods. 2006;3(2):95–7. https://doi.org/10.1038/nmeth850.
Chen J, Xu X, Huang Z, Luo Y, Tang L, Jiang JH. BEAMing LAMP: single-molecule capture and on-bead isothermal amplification for digital detection of hepatitis C virus in plasma. Chem Commun. 2018;54(3):291–4. https://doi.org/10.1039/C7CC08403J.
Shim J-u, Ranasinghe RT, Smith CA, Ibrahim SM, Hollfelder F, Huck WTS, Klenerman D, Abell C. Ultrarapid generation of femtoliter microfluidic droplets for single-molecule-counting immunoassays. ACS Nano. 2013;7(7):5955–64. https://doi.org/10.1021/nn401661d.
Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, An T, Yao S, Zheng L. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018;18(7):4226–32. https://doi.org/10.1021/acs.nanolett.8b01184.
Yelleswarapu V, Buser JR, Haber M, Baron J, Inapuri E, Issadore D. Mobile platform for rapid sub-picogram-per-milliliter, multiplexed, digital droplet detection of proteins. Proc Natl Acad Sci USA. 2019;116(10):4489–95. https://doi.org/10.1073/pnas.1814110116.
Gorris HH, Walt DR. Analytical chemistry on the femtoliter scale. Angew Chem Int Ed. 2010;49(23):3880–95. https://doi.org/10.1002/anie.200906417.
Leirs K, Dal Dosso F, Perez-Ruiz E, Decrop D, Cops R, Huff J, Hayden M, Collier N, Yu KXZ, Brown S, Lammertyn J. Bridging the gap between digital assays and point-of-care testing: automated, low cost, and ultrasensitive detection of thyroid stimulating hormone. Anal Chem. 2022;94(25):8919–27. https://doi.org/10.1021/acs.analchem.2c00480.
Rissin DM, Walt DR. Digital concentration readout of single enzyme molecules using femtoliter arrays and Poisson statistics. Nano Lett. 2006;6(3):520–3. https://doi.org/10.1021/nl060227d.
Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595–9. https://doi.org/10.1038/nbt.1641.
Warren AD, Gaylord ST, Ngan KC, Dumont Milutinovic M, Kwong GA, Bhatia SN, Walt DR. Disease detection by ultrasensitive quantification of microdosed synthetic urinary biomarkers. J Am Chem Soc. 2014;136(39):13709–14. https://doi.org/10.1021/ja505676h.
Wang X, Cohen L, Wang J, Walt DR. Competitive immunoassays for the detection of small molecules using single molecule arrays. J Am Chem Soc. 2018;140(51):18132–9. https://doi.org/10.1021/jacs.8b11185.
Cohen L, Hartman MR, Amardey-Wellington A, Walt DR. Digital direct detection of microRNAs using single molecule arrays. Nucleic Acids Res. 2017;45(14):e137. https://doi.org/10.1093/nar/gkx542.
Wang X, Walt DR. Simultaneous detection of small molecules, proteins and microRNAs using single molecule arrays. Chem Sci. 2020;11(30):7896–903. https://doi.org/10.1039/D0SC02552F.
Ter-Ovanesyan D, Gilboa T, Lazarovits R, Rosenthal A, Yu X, Li JZ, Church GM, Walt DR. Ultrasensitive measurement of both SARS-CoV-2 RNA and antibodies from saliva. Anal Chem. 2021;93(13):5365–70. https://doi.org/10.1021/acs.analchem.1c00515.
Gilboa T, Cohen L, Cheng C-A, Lazarovits R, Uwamanzu-Nna A, Han I, Griswold K Jr, Barry N, Thompson DB, Kohman RE, Woolley AE, Karlson EW, Walt DR. A SARS-CoV-2 neutralization assay using single molecule arrays. Angew Chem Int Ed. 2021;60(49):25966–72. https://doi.org/10.1002/anie.202110702.
Lam L, Iino R, Tabata KV, Noji H. Highly sensitive restriction enzyme assay and analysis: a review. Anal Bioanal Chem. 2008;391(7):2423–32. https://doi.org/10.1007/s00216-008-2099-4.
Rondelez Y, Tresset G, Tabata KV, Arata H, Fujita H, Takeuchi S, Noji H. Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat Biotechnol. 2005;23(3):361–5. https://doi.org/10.1038/nbt1072.
Lam L, Sakakihara S, Ishizuka K, Takeuchi S, Noji H. An integrated system for enzymatic cleavage and electrostretching of freely-suspended single DNA molecules. Lab Chip. 2007;7(12):1738–45. https://doi.org/10.1039/B711826K.
Sakakihara S, Araki S, Iino R, Noji H. A single-molecule enzymatic assay in a directly accessible femtoliter droplet array. Lab Chip. 2010;10(24):3355–62. https://doi.org/10.1039/C0LC00062K.
Kim SH, Iwai S, Araki S, Sakakihara S, Iino R, Noji H. Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip. 2012;12(23):4986–91. https://doi.org/10.1039/C2LC40632B.
Obayashi Y, Iino R, Noji H. A single-molecule digital enzyme assay using alkaline phosphatase with a cumarin-based fluorogenic substrate. Analyst. 2015;140(15):5065–73. https://doi.org/10.1039/C5AN00714C.
Ueno H, Kato M, Minagawa Y, Hirose Y, Noji H. Elucidation and control of low and high active populations of alkaline phosphatase molecules for quantitative digital bioassay. Protein Sci. 2021;30(8):1628–39. https://doi.org/10.1002/pro.4102.
Honda S, Minagawa Y, Noji H, Tabata KV. Multidimensional digital bioassay platform based on an air-sealed femtoliter reactor array device. Anal Chem. 2021;93(13):5494–502. https://doi.org/10.1021/acs.analchem.0c05360.
Cohen L, Cui N, Cai Y, Garden PM, Li X, Weitz DA, Walt DR. Single molecule protein detection with attomolar sensitivity using droplet digital enzyme-linked immunosorbent assay. ACS Nano. 2020;14(8):9491–501. https://doi.org/10.1021/acsnano.0c02378.
Wu C, Garden PM, Walt DR. Ultrasensitive detection of attomolar protein concentrations by dropcast single molecule assays. J Am Chem Soc. 2020;142(28):12314–23. https://doi.org/10.1021/jacs.0c04331.
Xu S, Wu J, Chen C, Zhang J, Wang Y, Xu H. A micro-chamber free digital biodetection method via the “sphere-labeled-sphere” strategy. Sens Actuators B Chem. 2021;337(15):129794. https://doi.org/10.1016/j.snb.2021.129794.
Akama K, Shirai K, Suzuki S. Droplet-free digital enzyme-linked immunosorbent assay based on a tyramide signal amplification system. Anal Chem. 2016;88(14):7123–9. https://doi.org/10.1021/acs.analchem.6b01148.
Fan W, Qi Y, Lu X, Ren W, Liu C, Li Z. An emulsion-free digital flow cytometric platform for the precise quantification of microRNA based on single molecule extension-illuminated microbeads (dFlowSeim). Chem Commun. 2020;56(52):7179–82. https://doi.org/10.1039/D0CC03059G.
Gao X, Teng X, Dai Y, Li J. Rolling circle amplification-assisted flow cytometry approach for simultaneous profiling of exosomal surface proteins. ACS Sens. 2021;6(10):3611–20. https://doi.org/10.1021/acssensors.1c01163.
Wu C, Dougan TJ. Walt DR. High-throughput, high-multiplex digital protein detection with attomolar sensitivity. ACS Nano. 2022;16(1):1025-1035. https://doi.org/10.1021/acsnano.1c08675.
Qi Y, Zhai Y, Fan W, Ren W, Li Z, Liu C. Click chemistry-actuated digital DNA walker confined on a single particle toward absolute microRNA quantification. Anal Chem. 2021;93(3):1620–6. https://doi.org/10.1021/acs.analchem.0c04073.
Zhang L, Fan W, Jia D, Feng Q, Ren W, Liu C. Microchamber-free digital flow cytometric analysis of T4 polynucleotide kinase phosphatase based on single-enzyme-to-single-bead space-confined reaction. Anal Chem. 2021;93(44):14828–36. https://doi.org/10.1021/acs.analchem.1c03724.
Li J, Wuethrich A, Sina AAI, Cheng HH, Wang Y, Behren A, Mainwaring PN, Trau M. A digital single-molecule nanopillar SERS platform for predicting and monitoring immune toxicities in immunotherapy. Nat Commun. 2021;12(1):1087. https://doi.org/10.1038/s41467-021-21431-w.
Cao L, Cui X, Hu J, Li Z, Choi JR, Yang Q, Lin M, Li Y, Xu F. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron. 2017;90(15):459–74. https://doi.org/10.1016/j.bios.2016.09.082.
Xie XS, Dunn RC. Probing single molecule dynamics. Science. 1994;265(5170):361–4. https://doi.org/10.1126/science.265.5170.361.
Demanelis K, Jasmine F, Chen LS, Chernoff M, Tong L, Delgado D, Zhang C, Shinkle J, Sabarinathan M, Lin H, et al. Determinants of telomere length across human tissues. Science. 2020;369(6509):eaaz6876. https://doi.org/10.1126/science.aaz6876.
Gao K, Zhou Y, Lu Q, Lu J, Su L, Su R, Zhang M, Tian Y, Wu L, Yan X. High-throughput human telomere length analysis at the single-chromosome level by FISH coupled with nano-flow cytometry. Anal Chem. 2021;93(27):9531–40. https://doi.org/10.1021/acs.analchem.1c01544.
Sakamoto S, Komatsu T, Watanabe R, Zhang Y, Inoue T, Kawaguchi M, Nakagawa H, Ueno T, Okusaka T, Honda K, Noji H, Urano Y. Multiplexed single-molecule enzyme activity analysis for counting disease-related proteins in biological samples. Sci Adv. 2020;6(11):eaay0888. https://doi.org/10.1126/sciadv.aay0888.
Luo Y, Viswanathan R, Hande MP, Loh AHP, Cheow LF. Massively parallel single-molecule telomere length measurement with digital real-time PCR. Sci Adv. 2020;6(34):eabb7944. https://doi.org/10.1126/sciadv.abb7944.
Funding
This work was supported by the National Natural Science Foundation of China (22074088 and 21622507), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R43), the Innovation Capability Support Program of Shaanxi (No. 2021TD-42), and the Fundamental Research Funds for the Central Universities (GK202101001 and GK202201009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, W., Ren, W. & Liu, C. Advances in optical counting and imaging of micro/nano single-entity reactors for biomolecular analysis. Anal Bioanal Chem 415, 97–117 (2023). https://doi.org/10.1007/s00216-022-04395-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04395-8