Abstract
This work presents an optimized gas chromatography–electron ionization–high-resolution mass spectrometry (GC-EI-HRMS) screening method. Different method parameters affecting data processing with the Agilent Unknowns Analysis SureMass deconvolution software were optimized in order to achieve the best compromise between false positives and false negatives. To this end, an accurate-mass library of 26 model compounds was created. Then, five replicates of mussel extracts were spiked with a mixture of these 26 compounds at two concentration levels (10 and 100 ng/g dry weight in mussel, 50 and 500 ng/mL in extract) and injected in the GC-EI-HRMS system. The results of these experiments showed that accurate mass tolerance and pure weight factor (combination of reverse-forward library search) are the most critical factors. The validation of the developed method afforded screening detection limits in the 2.5–5 ng range for passive sampler extracts and 1–2 ng/g for mussel sample extracts, and limits of quantification in the 0.6–3.2 ng and 0.1–1.8 ng/g range, for the same type of samples, respectively, for 17 model analytes. Once the method was optimized, an accurate-mass HRMS library, containing retention indexes, with ca. 355 spectra of derivatized and non-derivatized compounds was generated. This library (freely available at https://doi.org/10.5281/zenodo.5647960), together with a modified Agilent Pesticides Library of over 800 compounds, was applied to the screening of passive samplers, both of polydimethylsiloxane and polar chemical integrative samplers (POCIS), and mussel samples collected in Galicia (NW Spain), where a total of 75 chemicals could be identified.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sustainable development is seriously impaired by the amount of many different chemicals which are emitted into our environment. Thus, chemical pollution has been considered one of the planetary boundaries, and closely linked to another one (freshwater, since the availability of safe freshwater resources is seriously threatened by chemical pollution), where action is needed [1,2,3,4]. In this context, we need efficient analytical methods, such as high-resolution mass spectrometry (HRMS)–based screening methods, capable of providing information on a broad range of chemical pollutants [5, 6]. Such screening methods can be used, for instance, as a first step in prioritizing the compounds that could later be determined at a quantitative level and in order to provide a broader picture of the chemical pollution space. In this context, liquid chromatography–HRMS (LC-HRMS) has become the most prominent technique in environmental screening analysis [7,8,9,10,11,12,13,14]. However, LC-HRMS has a limited applicability for several compounds which exhibit poor ionization efficiency by electrospray. Atmospheric pressure chemical ionization (APCI) and atmospheric pressure photoionization interfaces have been shown to be able to expand the capabilities of LC-(HR)MS to more hydrophobic compounds [15,16,17], but, so far, have found limited use in HRMS screening studies. Hence, gas chromatography–HRMS (GC-HRMS) is a good complement to LC-HRMS, having as further advantages the fact of being an ideal technique for volatile analytes, its higher separation power, and being less prone to matrix effects. An additional advantage of GC is that linear retention indexes (RI) are already available for several chemicals, further providing confirmatory confidence of compounds identity [18].
Although some researchers proposed a screening workflow in GC-HRMS systems equipped with an APCI source very similar to LC-HRMS, this is typically performed by deconvolution of chromatograms obtained by (both low resolution and HRMS) GC–MS instruments equipped with classical electron impact (EI) sources. Deconvoluted spectra can be compared to libraries with a high probability to obtain a good match as compared with non-deconvoluted spectra. The NIST MS library is by far the most frequently employed one in GC–MS screening studies [19,20,21,22,23]. However, it has the inconvenience of containing low-resolution mass spectra. Moreover, conversely to LC–MS, the availability of accurate-mass HRMS libraries for GC-EI-MS is very limited.
Deconvolution of GC-HRMS data processing has evolved significatively due to the introduction of new powerful, both commercial (e.g., Agilent MassHunter Unknowns Analysis) and open-source (e.g., MZmine2), software. Several papers have been published in order to improve the data mining workflow using open-source software such as MZmine2 [24,25,26]. However, poor information is available dealing with commercial software workflows, such as Agilent MassHunter Unknowns Analysis. For a specific chromatogram, the deconvolution algorithm (of this last software) can generate hundreds or even thousands of detected peaks. Therefore, it is important to appropriately select the deconvolution parameters in order to minimize the number of false positives and negatives. In the literature, there are a limited number of screening studies published that describe the use of the Unknowns Analysis software for data treatment, e.g., [20, 27, 28], and only one of them optimized a single parameter (the narrow extracted mass window (EMW)) [27] affecting the identification process of the deconvolution algorithm. EMW is not currently used in the most recent versions of this software, as the deconvolution algorithm was completely redesigned to work with HRMS data acquired in profile mode.
The main goals of this study were the optimization of a GC-HRMS deconvolution workflow algorithm (Agilent Unknowns Analysis) and its application to coastal mussel samples and coastal/marine- and freshwater-deployed passive samplers. To this end, first of all, an EI-HRMS library of 26 model compounds (Table S1) was created and employed in order to find out the best operating conditions in terms of false positive/false negative balance with spiked marine biota samples (mussel) extracts. Once the method was optimized, an accurate-mass HRMS library, totalling 356 different spectra, including derivatized and non-derivatized compounds, was generated and employed, together with an Agilent EI-HRMS (modified) library of pesticides to a set of environmental samples. Such samples included passive samplers and mussels, because of their ability to capture chemical pollutants along time. Passive samplers, including, among others, polar organic chemical integrative samplers (POCIS) and polymeric materials like low-density polyethylene (LDPE) and polydimethylsiloxane (PDMS) [13, 29,30,31,32], have been successfully used for water monitoring, including screening studies. Bivalve molluscs, such as mussels, are an important filter feeding organisms which have been used as bioindicators in environmental monitoring programs [33,34,35] due to their capacity to accumulate contaminants.
Materials and methods
Reagents and sorbents
Dichloromethane (DCM) for pesticide residue analysis was provided by VWR Chemicals (Radnor, PA, USA). Acetonitrile (ACN) hypergrade for LC–MS, ethyl acetate (EtOAc) for GC–MS, and isooctane for GC were provided by Merck (Darmstadt, Germany). Sulfuric acid (H2SO4) 95–98% was provided by Panreac (Barcelona, Spain). The silylation reagent, N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA), and the C7–C40 saturated alkanes standard solution were provided by Sigma-Aldrich (San Louis, MO, USA). Methanol optima LC–MS grade was provided by Thermo Fisher Scientific (Waltham, MA, USA). Ultrapure water was obtained in the laboratory by purifying demineralized water in a Milli-Q Gradient A-10 system (Merck-Millipore, Bedford, MA, USA).
Florisil (60–100 mesh) and primary-secondary amine–bonded silica (PSA) were provided by Supelco (Bellefonte, PA, USA). Silica gel 60 (0.040–0.063 mm) was provided by Merck. Oasis HLB 12 cc (500 mg) cartridges were supplied by Waters (Milford, MA, USA). Polyethersulfone (PES) membranes Suport®-450 47 mm 0.45 µm were obtained by Pall Corporation (Ann Arbor, MI, USA). Polydimethylsiloxane (PDMS) elastomer with 2 mm diameter was from Goodfellow (Huntingdon, UK). Florisil and silica were washed in a PLE system using an ASE 200 (Dionex, Idstein, Germany) apparatus, equipped with 33 mL stainless steel extraction cells, using first acetonitrile and then ethyl acetate at 60 °C, and then dried into the oven at 120 °C for 24 h, in order to minimize blank contamination issues [36].
POCIS stainless steel holders were constructed by Nodosafer (Pontevedra, Spain) with 70 mm external diameter and 40 mm internal diameter. Before POCIS assembly, stainless steel material was washed with soap, Milli-Q water, and methanol. PES membranes were sonicated in methanol and PDMS elastomer in ethyl acetate three times each one (10 min each time) in an ultrasonic bath and dried at room temperature. The HLB sorbent was obtained from Oasis HLB 12 cc cartridges. Oasis HLB cartridges were conditioned with 20 mL of methanol and dried under nitrogen stream before being dismantled. POCIS was assembled enclosing 100 mg of HLB sorbent between two PES membranes. The sandwich was sealed with two stainless steel rings and bolted with three stainless steel screws [13].
Pieces of 50 cm of PDMS were cut and rolled around one stainless steel ring. POCIS devices and PDMS were placed in stainless steel cages with holes that let water run through and protect them against potential damages on the sampling points.
Sampling and deployment of passive samplers
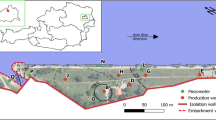
Mussel (Mytilus galloprovincialis) samples were collected on the Atlantic estuaries Ría de Arousa (coded A1-A3) and Ría de Vigo (coded V1-V3) located in the northern coast of Spain in 2019 (Figure S1). Mussels were homogenized and freeze-dried in amber glass bottles by the Galician Technological Institute for the Monitoring of the Marine Environment (INTECMAR) and sent to the University of Santiago de Compostela for analysis.
Steel canisters with the passive samplers were placed in 3 locations in rivers (coded R1-R3) and 4 locations in the estuary Ría de Arousa (coded S1-S4) from Galicia (northwest of Spain) during 2019 (Figure S1) for 1 and 2 weeks, respectively.
Sample treatment
Passive samplers’ desorption
At the end of the sampling period, passive samplers were collected and transported to the lab. After reception, POCIS and PDMS were cleaned with abundant Milli-Q water, wrapped in aluminum foil, and stored at − 20 °C until desorption.
POCIS were disassembled and particles of HLB sorbent were transferred into an empty SPE cartridge and packed between two frits. Compounds adsorbed in the POCIS sorbent were eluted by gravity using 10 mL of methanol [13].
PDMS desorption was carried out with 15 mL of ethyl acetate in a vial by shaking at 175 rpm for 60 min using an orbital shaker supplied by Science Basic Solutions (SBS) (Rubí, Spain) [37].
POCIS and PDMS extracts were concentrated to ca. 0.5 mL in a Turbovap II concentrator (Zymark, Hopkinton, MA), then evaporated to dryness under a purified nitrogen stream and finally reconstituted in 500 µL and 1000 µL of ethyl acetate, respectively. An aliquot of 75 µL of each extract was derivatized with 25% of MSTFA by heating in the oven for 60 min at 65 °C [38].
With each set of samples, procedural blanks of POCIS and PDMS were performed and submitted to the corresponding protocols.
Mussel sample extraction
Freeze-dried mussel samples were processed using two different matrix solid-phase dispersion (MSPD) methods, as to cover analytes with different properties. In the first one (method A), based on [39], 0.5 g of freeze-dried mussel sample was mixed into a glass mortar with 0.5 g of PSA. The homogenized mixture was transferred into a cartridge containing different cleanup sorbents (0.5 g of silica followed by 1.75 g of acidified silica (10% (w/w) H2SO4) and 1.75 g of Florisil (deactivated with 5% (w/w) H20). Analytes were eluted with 10 mL of dichloromethane. The eluate was concentrated to dryness under a nitrogen stream and reconstituted in 200 µL of isooctane.
In the second method (method B), based on [36], 0.5 g freeze-dried mussel was mixed with 1.2 g activated silica into a glass mortar. The homogeneous mixture was transferred into a cartridge containing 3 g of deactivated (5% (w/w) H2O) Florisil. Analytes were eluted with 10 mL of acetonitrile. The extract was concentrated into a Turbovap II concentrator and evaporated to dryness under a purified nitrogen stream and reconstituted in 100 µL of ethyl acetate. An aliquot of 50 µL of each extract obtained by method B was derivatized with 25% of the silylating reagent MSTFA by heating in an oven for 60 min at 65 °C [38].
Procedural blanks of both MSPD methods were performed with each batch of samples.
Gas chromatography–high-resolution mass spectrometry
A GC-HRMS system comprised of a 7890A gas chromatograph, a 7638B automatic sampler, and a 7200 quadrupole time-of-flight (QTOF) mass spectrometer from Agilent Technologies (Wilmington, DE, USA) was employed. The system was controlled by MassHunter Acquisition B.07.06 software (Agilent).
Chromatographic separation was carried out on an HP-5MS capillary column (30 m × 250 µm i.d., 0.25 µm film thickness) supplied by Agilent Technologies. High-purity helium (99.9999%, Nippon Gases, Madrid, Spain) was used as carrier gas at a constant flow rate of 1 mL/min. The temperatures of the transfer line, quadrupole, and source were set at 280 °C, 150 °C, and 230 °C, respectively. Oven temperature was programmed as follows: 50 °C (held for 1 min) ramped at 10 °C/min to 290 °C (held for 15 min). The total run time was 40 min and the solvent delay 3.5 min. Injections of 1 µL were made at 280 °C in splitless mode for 1 min using a 10 µL syringe. The injector was equipped with an Agilent ultra-inert liner containing glass wool.
The QTOF mass spectrometer was operated at 2-GHz in the EI mode at 70 eV with the emission current filament set at 5 μA and in single MS mode. Data was acquired in both centroid and profile mode in the range from 40 to 1000 m/z, at a frequency of 5 spectra/s [27] and providing a full width at half maximum (FWHM) resolution of ca. 5700 at 68 m/z and ca. 8400 at 502 m/z. The QTOF mass axis was automatically recalibrated every 3 injections by infusion of a commercial solution of perfluorotributylamine in the EI source.
Data analysis and in-house libraries for suspect screening
Data analysis was carried out with the Agilent MassHunter Unknowns Analysis B.10.00 software using the SureMass algorithm. Features were extracted by spectral deconvolution and the spectra for each feature were compared with those of a HRMS spectral library. In addition, retention indexes (RI) were also employed. Deconvolution, identification, and library search optimal parameters are described in Table 1.
For the analysis of environmental samples, two HRMS libraries were used: a commercial Agilent RTL Pesticides Library (modified to include RIs, see below) and the in-house library. A total of 300 compounds were selected for the creation of the in-house library. Standard solutions of ca. 1000 ng/mL were injected along with a C7–C40 alkane mix standard under the same chromatographic conditions, to obtain the spectra of individual compounds and calculate the corresponding Kovats RI. For 239 compounds, acceptable peak shape, retention, and intensity were obtained. Detailed information on name, CAS number, RI, and class from these individual contaminants is reported in Table S2. Moreover, for those compounds with reactive functional groups, an aliquot of standard was derivatized with a 25% of MSTFA by heating in an oven for 60 min at 65 °C [38]. Table S3 contains the 116 derivatized compounds, among which a non-derivatized spectrum was also available for 55 chemicals, while 61 of them produced a chromatographic peak only when derivatized. These 61 compounds include 26 pharmaceuticals, 20 human metabolites, 8 pesticides, 4 additives, and 3 transformation products. This library covers organic chemicals with molecular weights ranging from 104 to 692 Da, log Kow from − 1.2 to 9.6 and RIs from 900.5 to 3257.
The commercial library Agilent Pesticide AMRT PDCL (Table S4), which contains retention time (RT) values (when using a particular set of column and temperature program), was modified by including RIs. To this end, 12 chemicals with a wide range of retention times (RIs ranging from 1192 to 3257) and the n-alkanes mix were injected in the GC-QTOF with the Agilent reference oven program. Library retention time values were then updated using a linear correction between the library and the experimentally obtained retention times, and then converted into RIs.
A calibration retention time (CRT) file was generated for each set of samples using the C7–C40 n-alkane standard mix injected under the same chromatographic conditions. The CRT file is a csv file that contains the name, retention time, and the Kovats RI of each alkane and is used by the Unknowns Analysis software for RI matching.
Optimization of Unknowns Analysis deconvolution parameters
Several parameters of the Unknown Analysis method using the SureMass algorithm were optimized in order to achieve the best compromise between false positives and false negatives. To this end, a set of 26 model different compounds (see Table S1) was employed for the creation of an EI-HRMS library by injection of individual standards. These chemicals belong to different families (fragrances, plasticizers, phthalates, polycyclic aromatic hydrocarbons (PAHs), parabens, polybrominated diphenyl ethers (PBDEs), benzothiazoles, pesticides, polychlorinated biphenyls (PCBs), organophosphate esters, and UV filter compounds), with molecular weights between 128 and 481 Da and log Kow between 1.7 and 8.0. It also contains compounds with poor (in terms of number of ions) spectra (e.g., dibutyl phthalate, pyrene, and benzothiazole) and rich spectra (e.g., tris(2-butoxyethyl)phosphate, 4-methyl-1H-benzotriazole and triclosan), and a retention time range between 6.3 min (2-chlorophenol) and 26.3 min (benzo[a]pyrene), chemicals with and without Cl/Br atoms, and compounds showing a good peak shape (e.g., BHT) or tailing peaks (e.g., 2-chlorophenol).
Five replicates of mussel extracts (obtained by method B, as detailed above) were spiked with a mixture of these 26 compounds at two concentration levels (10 and 100 ng/g dry weight (dw) in mussel, equivalent to 50 ng/mL and 500 ng/mL in extract) and injected in the GC-QTOF system. Compound identification was performed by considering different method parameters (accurate mass tolerance (AMT), match factor, and pure weight factor (PWF)). From these parameters, AMT represents the maximum allowed difference between target and library exact masses; the score represents the cutoff value (100% is a perfect match) for positive identification, and PWF controls the forward and reverse matching balance, where the values of 0 and 1 represent a purely reverse and forward search, respectively, and the values in between a weighted combination of those extreme situations. AMT and match factor were first assessed at the highest concentration level in terms of false negatives, while the most critical parameter, PWF, was then optimized at the lowest concentration, where both false positives and negatives are considered. Retention times and RIs were not considered at this stage since we wanted to select the best conditions purely in spectral matching terms.
Method performance and quantitative analysis
From the compounds found in both water and mussel samples, 17 chemicals, whose standards were available in the lab, were selected for method performance evaluation. Mussel samples with low contamination levels and passive sampler sorbents were spiked with the selected compounds at two concentration levels (5 and 50 ng/mL referred to the extract, equivalent to 1–2 and 10–20 ng/g dw, respectively, depending on the MSPD method), analyzed (five replicates) together with the respective non-spiked samples and blanks according to the described procedures, submitted to the GC-EI-HRMS optimized screening workflow, and, finally, the percentage of chemicals positively detected was calculated. Then, the screening detection limit (SDL) was established as the lowest concentration for which it has been demonstrated that an analyte can be detected in at least 95% of the samples [40].
Trueness and LOQ evaluation for these 17 chemicals was performed by spiking passive sampler extracts or mussel samples. Trueness (recovery and repeatability) was evaluated at one concentration level (50 ng/mL referred to standard, equivalent 10–20 ng/g dw when referring to mussel samples). An estimation of their concentration in the samples was then performed by the external standard calibration method (LOQ-100 ng/mL calibration range).
Results and discussion
Optimization of the deconvolution parameters
Accurate mass tolerance
As a preliminary step, we optimized the AMT, which represents the maximum mass error (in ppm) allowed by the software during comparison of deconvoluted and library spectra. Thus, the samples spiked at 500 ng/mL (100 ng/g dw referred to sample) were processed with the Unknowns Analysis SureMass deconvolution algorithm with three different AMT values (10 ppm (default value), 20 ppm, and 50 ppm) combined to five different values of PWF (0, 0.25, 0.5, 0.75, and 1) and setting the minimum match factor to 50 (this value, in percentage, refers to spectral match against the library). Figure S2 shows the percentage of false negatives (compounds not being detected) in each combination of AMT and PWF. As observed, no compounds were identified at all using the default value of 10 ppm at any PWF. By increasing the AMT value, the percentage of false negatives decreased, being in the range 43–50% at 20 ppm and 0% at 50 ppm for any PWF. Therefore, the AMT was fixed at this latest value of 50 ppm. This relatively large AMT value is mostly due to the fact that the software used for creation of the library and the SureMass algorithm used during deconvolution perform a different interpolation for the conversion of profile to centroid spectra. Besides, other factors related to this are the limited resolution of the GC-QTOF and the fact that EI spectra contain many low masses. It is expected that software upgrading and improved performance of the latest generation of GC-EI-QTOF systems will help into reducing the AMT in the near future.
Establishment of match factor cutoff values
Once the AMT was set, the minimum library match factor for the identification of the chemicals was evaluated at six different values of PWF (0, 0.1, 0.25, 0.5, 0.75, and 1) with the samples spiked at the highest concentration level (500 ng/mL referred to extract, 100 ng/g dw referred to sample). The distribution of match factor values obtained at the different PWFs is presented as Box-and-Whisker plots in Figure S3. Thus, the match factor cutoff values allowing a 0%, 1%, and 5% of false negatives could be established as the values presented in Table 2. As it can be observed, lower match factor cutoff values were required at higher PWFs (i.e., when forward search poses higher weights) for the positive identification of the analytes. The ideal experimental parameters should enable a 0% of false negatives but, unfortunately, this rarely happens. Hence, in agreement with the recommendation of the SANTE/12682/2019 guide [41] for screening methods, 5% false negatives cutoff values were used to further evaluate PWFs.
Selection of pure weight factor
As a final optimization step, the most critical parameter in terms of false positives and negatives was optimized at a lower concentration level. Hence, the samples spiked at 50 ng/mL referred to extract (10 ng/g dw referred to sample) were processed with different PWF values using the corresponding match factor cutoff for 5% of false negatives set at the highest spiked level (see above). A summary of false negatives (not detected) and positives (compounds that are incorrectly identified by the software) obtained during this final optimization step is displayed in Fig. 1. As it can be observed, the number of false positives decreases as the PWF moves from a pure reverse (PWF = 0) to a purely forward search (PWF = 1). Conversely, the false negatives rate, which is now higher than 5% due to the lowest spiked concentration, followed the opposite pattern, with PWF 0 and 0.1 leading to the lowest number of false negatives and PWF values in the 0.25–1 range leading to a higher rate of false negatives. Therefore, as a compromise, a PWF of 0.1, which provides the best balance between false positives and negatives at lower concentration levels, was selected as optimal. Although this would result on an expectable rate of false positives of ca. 30%, this value is in practice far lower when the RI matching is implemented.
Rate of false negatives and false positives as a function of the pure weight factor (PFW) for the 50 ng/mL spiked mussel extract (10 ng/g dw referring to sample). Match factor cutoff set to allow a 5% of false negatives of the 500 ng/mL level (100 ng/g dw referring to sample) (see Table 2)
Method performance
The proposed method performance was investigated with 17 compounds that were detected in the samples (see “Application to passive sampler and mussel samples”) at two concentration levels as explained in “Material and methods.” For some compounds found in the non-spiked samples at comparatively high concentration levels (3 and 5 compounds for MSPD methods A and B, respectively), the detection frequency could not be calculated in some cases (Table S5). As shown in Table S5, the detection frequency was 100% for most of the compounds at the two concentration levels for the four protocols. However, in the case of passive samplers, lower detection frequencies were observed for the most hydrophobic compounds (i.e., 2-ethylhexyl salicylate, triclosan, 2-ethylhexyl-4-methoxycinnamate, and pyrene) using POCIS. On the other hand, 2,6-di-tert-butyl-4-methylphenol (BHT) and triclosan presented lower detection frequencies in mussel samples likely due to the degradation with sulfuric acid in method A. From such data, SDLs were derived (Table 3).
The SDL for passive samplers was determined to be 5 ng for PDMS (the lowest level tested). In the case of the POCIS, this SDL was 2.5 ng (the lowest level tested) for most compounds, except for 2-ethylhexyl salicylate and pyrene (SDL: 25 ng), where triclosan could only be identified in the derivatized samples. Considering typical values of sampling rate (Rs) ranging between 0.01 and 1 L/day [42,43,44,45], that would translate into SDLs referring to water in the 0.18–35 ng/L range, depending on the actual Rs value and deployment time of the passive sampler (1 or 2 weeks). Reported SDL values in screening studies of surface waters, most of them based on LC-HRMS, were in the range 1.25–500 ng/L [46,47,48]. As regards mussel samples, not all chemicals’ SDL values could be calculated by both methods, either because of their degradation under the acidic treatment of method A (which targeted more stable compounds) or relatively high levels in the non-spiked sample. Otherwise, SDL values of 1 or 2 ng/g dw (considering an average humidity of 85% that would be equivalent to 0.15–0.30 ng/g wet weight (ww)) could be achieved for all the compounds using at least one of the proposed methods (Table 3). In the literature, SDL values can be found for vegetables, fruits [49,50,51], and feed but only few studies consider biota samples (fish) with SDL values of 5 ng/g ww [48].
Besides SDLs, recoveries and limits of quantification were also calculated (Table 4) in order to, later on, estimate the concentrations for those 17 chemicals whose standards were available in the environmental samples. The passive sampling and MSPD method used was the one where the chemicals were more often detected. Good absolute recoveries, between 82 and 107% in mussel samples and between 80 and 108% in passive samplers, and precision (RSD values < 9%) were obtained for most compounds (Table 4). LOQs in passive samplers ranged between 0.6 and 3.2 ng. Considering the above mentioned Rs values, when referring to real samples, LOQs would range between 0.04 and 46 ng/L. In the case of mussel samples, LOQs were in the 0.1–1.8 ng/g dw (equivalent to ca. 0.015–0.27 ng/g ww) range.
Application to passive sampler and mussel samples
The optimized screening method was applied to samples from the marine environment and river water from Galicia (NW Spain), including mussels and passive samplers. A total of 75 compounds were identified in these samples (summarized in Table 5, further details on each particular sample and compound usage is presented in Table S6), 46 of which were identified only in the passive samplers and 52 only in mussels, while 23 could be positively detected in both water and mussel samples. The use of RI allowed the identification of isomers or compounds with similar spectra, e.g., di-iso-butyl phthalate and di-n-butyl phthalate (Fig. 2). Further examples of some of deconvoluted spectra as compared to the libraries’ spectra are presented in Figure S5.
By sample (Fig. 3a), the number of compounds detected in passive samplers was in the range of 12 to 29, without clear differences between estuary and river water. This may be due to the longer deployment time of the passive samplers in the marine environment (1 vs 2 weeks in freshwater and marine water, respectively), which was performed as to compensate for the expectably lower concentrations. R2, in river, and S2, in estuarine, samples, which are the sampling points closer to the discharge of wastewater treatment plants (WWTPs), were the most polluted samples in terms of detected chemicals, with 23 and 29 identified compounds, respectively. As regards mussel samples (Fig. 3b), the number of identified compounds ranged from 9 to 32, being this number quite stable in Vigo estuary (29–32 compounds at each location) but more variable between different locations of Arousa estuary (9–26 compounds), where the sampling point A3 was far cleaner (Fig. 3b).
Regarding sampling mode, more compounds per sample were detected in PDMS than in POCIS, with 2–4 compounds detected in both samplers, except in Sample S2 where more compounds were identified in the POCIS (Fig. 3a). The different patterns in this sample are mostly due to the fact that polar compounds related with the proximity to a WWTP, such as the silylated derivatives of ibuprofen, naproxen, and ketoprofen or dimethyl phthalate, were detected here. Thus, these results show the complementarity of these two passive sampling techniques for the detection of contaminants. In mussel samples, the number of compounds detected by MSPD method A was lower than by method B, with 0–7 compounds detected by both methods, but for the cleanest sample, A3, where more compounds were detected by method A (Fig. 3b).
By library, 70% of the compounds detected in the passive samplers were non-derivatized analytes from the in-house library (55% in POCIS and 82% in PDMS), 11% from the pesticides library (21% in POCIS and 9% in PDMS), 13% derivatized compounds from the in-house library (21% in POCIS and 9% in PDMS), and the rest (7%) from the two libraries (Figure S4b). A similar distribution was found in mussel samples, i.e., 77% from non-derivatized analytes (in-house library), 12% from pesticides library, 4% of derivatized compounds (in-house library), and 8% from two of the libraries (Figure S4a). These findings highlight the relevance of expanding high-resolution EI-MS libraries.
Plasticizers were the major contaminants detected in all the environmental samples, which collectively accounted for 25–38% of the total number of contaminants for each matrix. In particular, 4 plasticizers, i.e., dicyclohexyl phthalate, di-iso-butyl phthalate, diethylphthalate, and di-n-butyl phthalate, were the compounds more frequently detected in all samples (> 85% samples). However, in water, the type of compounds detected in each passive sampler differs. While in POCIS, pharmaceuticals represent a 17% of the total number of compounds, being the second class of pollutants, and pesticides the third-class with a 14%; in PDMS, UV filters, fragrances, and pesticides represent the second class of chemicals with a 12% detection rate each. In mussels, 83% of the compounds detected belong to one of the following classes: plasticizers (23%), industrial chemicals (23%), pesticides (17%), PAHs (10%), and fragrances (10%). The type of compounds detected also differs by the MSPD method used. From the most frequently detected families, plasticizers are detected by both methods, pesticides and fragrances are mainly detected in MSPD method B extracts, while PAHs in MSPD method A extracts. On the other hand, industrial chemicals, which include compounds with different chemical characteristics, are detected, depending on the structure, by method A (e.g., trichlorobenzenes) or by method B (e.g., phenols). Finally, it is clear that mussels and PDMS can capture more hydrophobic chemicals as compared to POCIS, which are designed to detect more polar analytes.
Estimated concentrations
The concentrations (Table 6) of the 17 compounds detected in the samples, which could be validated (see “Method performance”), were calculated using the method selected in Table 4. In passive samplers, different profiles were observed. Thus, on the one hand, some compounds (e.g., diethyl phthalate and galaxolide) were found at higher concentration in passive samplers close to sewage discharges, pointing to wastewater treatment plants as their main source, whereas, for instance, on the other hand, EHMC was detected only in marine samples. Considering the typical values of sampling rate (Rs) of 0.01 and 1 L/day [42,43,44,45], the amounts measured would represent concentrations in the sub-ng/L to low µg/L level.
In mussel samples, the plasticizers diethyl phthalate and di-n-butyl phthalate and the UV filter EHMC were the compounds found at higher concentrations; up to 2746 ng/g dw for di-n-butyl phthalate (equivalent to 412 ng/g ww).
Conclusions
We have optimized the different parameters used for the GC-EI-HRMS screening of organic contaminants by the Agilent Unknowns Analysis SureMass algorithm, finding out that the selection of mass tolerance and weighting of reversed-/forward-searching plays a relevant role in the potential number of false positives and false negatives. Furthermore, an HRMS library was created, where the inclusion of RIs further provides confidence on the identification. The optimized method provides good SDL and was able to successfully identify 75 chemicals in marine and freshwater samples.
References
Li M, Wiedmann T, Fang K, Hadjikakou M. The role of planetary boundaries in assessing absolute environmental sustainability across scales. Environ Int. 2021;152:106475. https://doi.org/10.1016/j.envint.2021.106475.
Diamond ML, de Wit CA, Molander S, Scheringer M, Backhaus T, Lohmann R, Arvidsson R, Bergman Å, Hauschild M, Holoubek I, Persson L, Suzuki N, Vighi M, Zetzsch C. Exploring the planetary boundary for chemical pollution. Environ Int. 2015;78:8–15. https://doi.org/10.1016/j.envint.2015.02.001.
Wang Z, Altenburger R, Backhaus T, Covaci A, Diamond ML, Grimalt JO, Lohmann R, Schäffer A, Scheringer M, Selin H, Soehl A, Suzuki N. We need a global science-policy body on chemicals and waste. Science. 2021;80(371):774–6. https://doi.org/10.1126/science.abe9090.
István P (2020) The European environment-state and outlook 2020. Knowledge for transition to a sustainable Europe
Hernández F, Sancho JV, Ibáñez M, Abad E, Portolés T. Mattioli L (2012) Current use of high-resolution mass spectrometry in the environmental sciences. Anal Bioanal Chem. 2012;4035(403):1251–64. https://doi.org/10.1007/S00216-012-5844-7.
Lehotay SJ, Sapozhnikova Y, Mol HGJ. Current issues involving screening and identification of chemical contaminants in foods by mass spectrometry. TrAC - Trends Anal Chem. 2015;69:62–75. https://doi.org/10.1016/j.trac.2015.02.012.
Joye T, Sidibé J, Déglon J, Karmime A, Sporkert F, Widmer C, Favrat B, Lescuyer P, Augsburger M, Thomas A. Liquid chromatography-high resolution mass spectrometry for broad-spectrum drug screening of dried blood spot as microsampling procedure. Anal Chim Acta. 2019;1063:110–6. https://doi.org/10.1016/j.aca.2019.02.011.
Salgueiro-González N, Castiglioni S, Gracia-Lor E, Bijlsma L, Celma A, Bagnati R, Hernández F, Zuccato E. Flexible high resolution-mass spectrometry approach for screening new psychoactive substances in urban wastewater. Sci Total Environ. 2019;689:679–90. https://doi.org/10.1016/j.scitotenv.2019.06.336.
de Sardela PDO, Sardela VF, dos da Silva AMS, Pereira HMG, de Aquino Neto FR. A pilot study of non-targeted screening for stimulant misuse using high-resolution mass spectrometry. Forensic Toxicol. 2019;37:465–73. https://doi.org/10.1007/s11419-019-00482-1.
Meng D, Fan DL, Gu W, Wang Z, Chen YJ, Bu HZ, Liu JN. Development of an integral strategy for non-target and target analysis of site-specific potential contaminants in surface water: a case study of Dianshan Lake, China. Chemosphere. 2020;243:125367. https://doi.org/10.1016/j.chemosphere.2019.125367.
Wilson EW, Castro V, Chaves R, Espinosa M, Rodil R, Quintana JB, Vieira MN, Santos MM. Using zebrafish embryo bioassays combined with high-resolution mass spectrometry screening to assess ecotoxicological water bodies quality status: a case study in Panama rivers. Chemosphere. 2021;272:129823. https://doi.org/10.1016/j.chemosphere.2021.129823.
Montes R, Aguirre J, Vidal X, Rodil R, Cela R, Quintana JB. Screening for polar chemicals in water by trifunctional mixed-mode liquid chromatography-high resolution mass spectrometry. Environ Sci Technol. 2017;51:6250–9. https://doi.org/10.1021/acs.est.6b05135.
Castro V, Quintana JB, Carpinteiro I, Cobas J, Carro N, Cela R, Rodil R. Combination of different chromatographic and sampling modes for high-resolution mass spectrometric screening of organic microcontaminants in water. Anal Bioanal Chem. 2021. https://doi.org/10.1007/s00216-021-03226-6.
Gago-Ferrero P, Schymanski EL, Bletsou AA, Aalizadeh R, Hollender J, Thomaidis NS. Extended suspect and non-target strategies to characterize emerging polar organic contaminants in raw wastewater with LC-HRMS/MS. Environ Sci Technol. 2015;49:12333–41. https://doi.org/10.1021/acs.est.5b03454.
Qu G, Shi J, Wang T, Fu J, Li Z, Wang P, Ruan T, Jiang G. Identification of tetrabromobisphenol a diallyl ether as an emerging neurotoxicant in environmental samples by bioassay-directed fractionation and HPLC-APCI-MS/MS. Environ Sci Technol. 2011;45:5009–16. https://doi.org/10.1021/es2005336.
Zhou SN, Reiner EJ, Marvin C, Helm P, Riddell N, Dorman F, Misselwitz M, Shen L, Crozier P, MacPherson K, Brindle ID. Development of liquid chromatography atmospheric pressure chemical ionization tandem mass spectrometry for analysis of halogenated flame retardants in wastewater. Anal Bioanal Chem. 2010;396:1311–20. https://doi.org/10.1007/s00216-009-3279-6.
Zacs D, Bartkevics V. Analytical capabilities of high performance liquid chromatography - atmospheric pressure photoionization - Orbitrap mass spectrometry (HPLC-APPI-Orbitrap-MS) for the trace determination of novel and emerging flame retardants in fish. Anal Chim Acta. 2015;898:60–72. https://doi.org/10.1016/j.aca.2015.10.008.
van Den Dool H, Dec Kratz P. A generalization of the retention index system including linear temperature programmed gas—liquid partition chromatography. J Chromatogr A. 1963;11:463–71. https://doi.org/10.1016/S0021-9673(01)80947-X.
Blum KM, Andersson PL, Ahrens L, Wiberg K, Haglund P. Persistence, mobility and bioavailability of emerging organic contaminants discharged from sewage treatment plants. Sci Total Environ. 2018;612:1532–42. https://doi.org/10.1016/j.scitotenv.2017.09.006.
Lee S, Kim K, Jeon J, Moon HB. Optimization of suspect and non-target analytical methods using GC/TOF for prioritization of emerging contaminants in the Arctic environment. Ecotoxicol Environ Saf. 2019;181:11–7. https://doi.org/10.1016/j.ecoenv.2019.05.070.
Cha J, Hong S, Kim J, Lee J, Yoon SJ, Lee S, Moon HB, Shin KH, Hur J, Giesy JP, Khim JS. Major AhR-active chemicals in sediments of Lake Sihwa, South Korea: application of effect-directed analysis combined with full-scan screening analysis. Environ Int. 2019;133:105199. https://doi.org/10.1016/j.envint.2019.105199.
Gómez-Ramos MM, Ucles S, Ferrer C, Fernández-Alba AR, Hernando MD. Exploration of environmental contaminants in honeybees using GC-TOF-MS and GC-Orbitrap-MS. Sci Total Environ. 2019;647:232–44. https://doi.org/10.1016/j.scitotenv.2018.08.009.
Hoh E, Dodder NG, Lehotay SJ, Pangallo KC, Reddy CM, Maruya KA. Nontargeted comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry method and software for inventorying persistent and bioaccumulative contaminants in marine environments. Environ Sci Technol. 2012;46:8001–8. https://doi.org/10.1021/es301139q.
Korf A, Hammann S, Schmid R, Froning M, Hayen H. Cramp LJE (2020) Digging deeper - a new data mining workflow for improved processing and interpretation of high resolution GC-Q-TOF MS data in archaeological research. Sci Reports. 2020;101(10):1–9. https://doi.org/10.1038/s41598-019-57154-8.
Smirnov A, Qiu Y, Jia W, Walker DI, Jones DP, Du X. ADAP-GC 4.0: Application of clustering-assisted multivariate curve resolution to spectral deconvolution of gas chromatography–mass spectrometry metabolomics data. Anal Chem. 2019;91:9069–77. https://doi.org/10.1021/acs.analchem.9B01424.
Pluskal T, Castillo S, Villar-Briones A. Orešič M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinforma. 2010;111(11):1–11. https://doi.org/10.1186/1471-2105-11-395.
Zhang F, Wang H, Zhang L, Zhang J, Fan R, Yu C, Wang W, Guo Y. Suspected-target pesticide screening using gas chromatography-quadrupole time-of-flight mass spectrometry with high resolution deconvolution and retention index/mass spectrum library. Talanta. 2014;128:156–63. https://doi.org/10.1016/j.talanta.2014.04.068.
Moschet C, Lew BM, Hasenbein S, Anumol T, Young TM. LC- and GC-QTOF-MS as complementary tools for a comprehensive micropollutant analysis in aquatic systems. Environ Sci Technol. 2017;51:1553–61. https://doi.org/10.1021/acs.est.6b05352.
Allan IJ, Harman C, Ranneklev SB, Thomas KV, Grung M. Passive sampling for target and nontarget analyses of moderately polar and nonpolar substances in water. Environ Toxicol Chem. 2013;32:1718–26. https://doi.org/10.1002/etc.2260.
Yates K, Davies I, Webster L, Pollard P, Lawton L, Moffat C. Passive sampling: partition coefficients for a silicone rubber reference phase. J Environ Monit. 2007;9:1116–21. https://doi.org/10.1039/b706716j.
Gravell A, Fones GR, Greenwood R, Mills GA. Detection of pharmaceuticals in wastewater effluents—a comparison of the performance of Chemcatcher® and polar organic compound integrative sampler. Environ Sci Pollut Res. 2020;27:27995–8005. https://doi.org/10.1007/s11356-020-09077-5.
Menger F, Ahrens L, Wiberg K, Gago-Ferrero P. Suspect screening based on market data of polar halogenated micropollutants in river water affected by wastewater. J Hazard Mater. 2021;401:123377. https://doi.org/10.1016/j.jhazmat.2020.123377.
Mariné Oliveira GF, do Couto MCM, de Freitas Lima M, do Bomfim TCB. Mussels (Perna perna) as bioindicator of environmental contamination by Cryptosporidium species with zoonotic potential. Int J Parasitol Parasites Wildl. 2016;5:28–33. https://doi.org/10.1016/j.ijppaw.2016.01.004.
Goto A, Tue NM, Isobe T, Takahashi S, Tanabe S, Kunisue T. Nontarget and target screening of organohalogen compounds in mussels and sediment from Hiroshima Bay, Japan: occurrence of novel bioaccumulative substances. Environ Sci Technol. 2020;54:5480–8. https://doi.org/10.1021/acs.est.9b06998.
Rodil R, Villaverde-de-Sáa E, Cobas J, Quintana JB, Cela R, Carro N. Legacy and emerging pollutants in marine bivalves from the Galician coast (NW Spain). Environ Int. 2019;129:364–75. https://doi.org/10.1016/j.envint.2019.05.018.
Castro V, Montes R, Quintana JB, Rodil R, Cela R. Determination of 18 organophosphorus flame retardants/plasticizers in mussel samples by matrix solid-phase dispersion combined to liquid chromatography-tandem mass spectrometry. Talanta. 2019;208:120470. https://doi.org/10.1016/j.talanta.2019.120470.
Prieto A, Rodil R, Quintana JB, Rodríguez I, Cela R, Möder M. Evaluation of low-cost disposable polymeric materials for sorptive extraction of organic pollutants in water samples. Anal Chim Acta. 2012;716:119–27. https://doi.org/10.1016/j.aca.2011.12.023.
Quintana JB, Carpinteiro J, Rodríguez I, Lorenzo RA, Carro AM, Cela R (2004) Determination of natural and synthetic estrogens in water by gas chromatography with mass spectrometric detection. J Chromatogr A. 2004;1024:177–185. https://doi.org/10.1016/j.chroma.2003.10.074.
Villaverde-de-Sáa E, Valls-Cantenys C, Quintana JB, Rodil R, Cela R. Matrix solid-phase dispersion combined with gas chromatography-mass spectrometry for the determination of fifteen halogenated flame retardants in mollusks. J Chromatogr A. 2013;1300:85–94. https://doi.org/10.1016/j.chroma.2013.05.064.
European Commission (2017) Guidance document on analytical quality control and method validation procedures for pesticides residues analysis in food and feed. SANTE/11813/2017.
European Commission (2019) Analytical quality control and method validation for pesticide residues analysis in food and feed (SANTE/12682/2019).
Vrana B, Komancová L, Sobotka J. Calibration of a passive sampler based on stir bar sorptive extraction for the monitoring of hydrophobic organic pollutants in water. Talanta. 2016;152:90–7. https://doi.org/10.1016/j.talanta.2016.01.040.
Męczykowska H, Kobylis P, Stepnowski P, Caban M. Calibration of passive samplers for the monitoring of pharmaceuticals in water-sampling rate variation. Crit Rev Anal Chem. 2017;47:204–22. https://doi.org/10.1080/10408347.2016.1259063.
Vrana B, Urík J, Fedorova G, Švecová H, Grabicová K, Golovko O, Randák T, Grabic R (2021) In situ calibration of polar organic chemical integrative sampler (POCIS) for monitoring of pharmaceuticals in surface waters. Environ Pollut. 269;2021:116121. https://doi.org/10.1016/j.envpol.2020.116121.
Martin A, Margoum C, Jolivet A, Assoumani A, El Moujahid B, Randon J, Coquery M. Calibration of silicone rubber rods as passive samplers for pesticides at two different flow velocities: modeling of sampling rates under water boundary layer and polymer control. Environ Toxicol Chem. 2018;37:1208–18. https://doi.org/10.1002/etc.4050.
Hernández F, Ibáñez M, Portolés T, Cervera MI, Sancho JV, López FJ. Advancing towards universal screening for organic pollutants in waters. J Hazard Mater. 2015;282:86–95. https://doi.org/10.1016/j.jhazmat.2014.08.006.
Gago-Ferrero P, Bletsou AA, Damalas DE, Aalizadeh R, Alygizakis NA, Singer HP, Hollender J, Thomaidis NS. Wide-scope target screening of >2000 emerging contaminants in wastewater samples with UPLC-Q-ToF-HRMS/MS and smart evaluation of its performance through the validation of 195 selected representative analytes. J Hazard Mater. 2020;387:121712. https://doi.org/10.1016/j.jhazmat.2019.121712.
Diamanti KS, Alygizakis NA, Nika MC, Oswaldova M, Oswald P, Thomaidis NS, Slobodnik J. Assessment of the chemical pollution status of the Dniester River Basin by wide-scope target and suspect screening using mass spectrometric techniques. Anal Bioanal Chem. 2020;412:4893–907. https://doi.org/10.1007/s00216-020-02648-y.
Mol HGJ, Zomer P, De Koning M. Qualitative aspects and validation of a screening method for pesticides in vegetables and fruits based on liquid chromatography coupled to full scan high resolution (Orbitrap) mass spectrometry. Anal Bioanal Chem. 2012;403:2891–908. https://doi.org/10.1007/s00216-012-6100-x.
Boix C, Ibáñez M, Sancho JV, León N, Yusá V, Hernández F. Qualitative screening of 116 veterinary drugs in feed by liquid chromatography-high resolution mass spectrometry: potential application to quantitative analysis. Food Chem. 2014;160:313–20. https://doi.org/10.1016/j.foodchem.2014.03.086.
Pang G, Chang Q, Bai R, Fan C, Zhang Z, Yan H, Wu X. Simultaneous screening of 733 pesticide residues in fruits and vegetables by a GC/LC-Q-TOFMS combination technique. Engineering. 2020;6:432–41. https://doi.org/10.1016/j.eng.2019.08.008.
Acknowledgements
The authors thank “Centro de Supercomputación de Galicia (CESGA)” for the use of their computational resources for HRMS data processing.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This research was funded by Xunta de Galicia (ED431C 2021/06 and V.C. predoctoral contract: ED481A-2017/156), the Spanish Agencia Estatal de Investigación—MCIN/AEI/10.13039/501100011033 (ref. CTM2017-84763-C3-R-2 and PID2020-117686RB-C32), and FEDER/ERDF funds.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Open access to the in-house libraries
The in-house libraries, in PCDL Agilent and JCAMP formats, are freely available at https://doi.org/10.5281/zenodo.5647960.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Published in the topical collection featuring Sustainability in (Bio-)Analytical Chemistry.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Castro, V., Quintana, J.B., López-Vázquez, J. et al. Development and application of an in-house library and workflow for gas chromatography–electron ionization–accurate-mass/high-resolution mass spectrometry screening of environmental samples. Anal Bioanal Chem 414, 6327–6340 (2022). https://doi.org/10.1007/s00216-021-03810-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03810-w