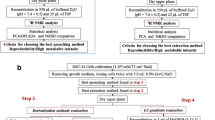
Abstract
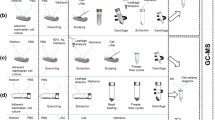
Cellular metabolomics has become key to elucidate mechanistic aspects in various fields such as cancerology or pharmacology, and is rapidly becoming a standard phenotyping tool accessible to the broad biological community. Acquisition of reliable spectroscopic datasets, such as nuclear magnetic resonance (NMR) spectra, to characterize biological systems depends on the elaboration of robust methods for cellular metabolites extraction. Previous studies have addressed many issues raised by these protocols, however with little pondering on ergonomic and practical aspects of the methods that impact their scalability, reproducibility and hence their suitability to high-throughput studies or their use by non-metabolomics experts. Here, we optimize a fast and ergonomic protocol for extraction of metabolites from adherent mammalian cells for NMR metabolomics studies. The proposed extraction protocol, including cell washing, metabolism quenching and actual extraction of intracellular metabolites, was first optimized on HeLa cells. Efficiency of the protocol, in its globality and for the different individual steps, was assessed by NMR quantification of 27 metabolites from cellular extracts. We show that a single PBS wash provides a seemly compromise between contamination from growth medium and leakage of intracellular metabolites. In HeLa cells, extraction using pure methanol, without cell scraping, recovered a higher amount of intracellular metabolites than the reference methanol/water/chloroform method with cell scraping, with yields varying across metabolite classes. Optimized and reference protocols were further tested on eight cell lines of miscellaneous nature, and inter-operator reproducibility was demonstrated. Our results stress the need for tailored extraction protocols and show that fast protocols minimizing time-consuming steps, without compromising extraction yields, are suitable for high-throughput metabolomics studies.

Graphical abstract





Similar content being viewed by others
References
Zhang A, Sun H, Wang P, et al. Recent and potential developments of biofluid analyses in metabolomics. J Proteome. 2012;75:1079–88.
Giskeødegård GF, Madssen TS, Euceda LR, et al. NMR-based metabolomics of biofluids in cancer. NMR Biomed. 2018;32:e3927.
Zhang A, Sun H, Xu H, et al. Cell metabolomics. OMICS. 2013;17:495–501.
Rinschen MM, Ivanisevic J, Giera M, et al. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol. 2019;20:353–67.
Aranibar N, Borys M, Mackin NA, et al. NMR-based metabolomics of mammalian cell and tissue cultures. J Biomol NMR. 2011;49:195–206.
Lefevre C, Panthu B, Naville D, et al. Metabolic phenotyping of adipose-derived stem cells reveals a unique signature and intrinsic differences between fat pads. Stem Cells Int. 2019;2019:1–16.
Moussaieff A, Rouleau M, Kitsberg D, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402.
Li H, Ning S, Ghandi M, et al. The landscape of cancer cell line metabolism. Nat Med. 2019;25:850–60.
Ortmayr K, Dubuis S, Zampieri M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism. Nat Commun. 2019;10:1841.
García-Cañaveras JC, Castell JV, Donato MT, et al. A metabolomics cell-based approach for anticipating and investigating drug-induced liver injury. Sci Rep. 2016;6:27239.
Hayton S, Maker GL, Mullaney I, et al. Experimental design and reporting standards for metabolomics studies of mammalian cell lines. Cell Mol Life Sci. 2017;74:4421–41.
Kapoore RV, Coyle R, Staton CA, et al. Influence of washing and quenching in profiling the metabolome of adherent mammalian cells: a case study with the metastatic breast cancer cell line MDA-MB-231. Analyst. 2017;142:2038–49.
Dettmer K, Nürnberger N, Kaspar H, et al. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal Bioanal Chem. 2011;399:1127–39.
Danielsson APH, Moritz T, Mulder H, et al. Development and optimization of a metabolomic method for analysis of adherent cell cultures. Anal Biochem. 2010;404:30–9.
Kapoore RV, Coyle R, Staton CA, et al. Cell line dependence of metabolite leakage in metabolome analyses of adherent normal and cancer cell lines. Metabolomics. 2015;11:1743–55.
Hutschenreuther A, Kiontke A, Birkenmeier G, et al. Comparison of extraction conditions and normalization approaches for cellular metabolomics of adherent growing cells with GC-MS. Anal Methods. 2012;4:1953.
Ser Z, Liu X, Tang NN, et al. Extraction parameters for metabolomics from cultured cells. Anal Biochem. 2015;475:22–8.
Lorenz MA, Burant CF, Kennedy RT. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal Chem. 2011;83:3406–14.
García-Cañaveras JC, López S, Castell JV, et al. Extending metabolome coverage for untargeted metabolite profiling of adherent cultured hepatic cells. Anal Bioanal Chem. 2016;408:1217–30.
Bi H, Krausz KW, Manna SK, et al. Optimization of harvesting, extraction, and analytical protocols for UPLC-ESI-MS-based metabolomic analysis of adherent mammalian cancer cells. Anal Bioanal Chem. 2013;405:5279–89.
Peterson A, Walker A, Sloan E, et al. Optimized method for untargeted metabolomics analysis of MDA-MB-231 breast cancer cells. Metabolites. 2016;6:30.
Teng Q, Huang W, Collette TW, et al. A direct cell quenching method for cell-culture based metabolomics. Metabolomics. 2009;5:199–208.
Martineau E, Tea I, Loaëc G, et al. Strategy for choosing extraction procedures for NMR-based metabolomic analysis of mammalian cells. Anal Bioanal Chem. 2011;401:2133–42.
Le Belle JE, Harris NG, Williams SR, et al. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed. 2002;15:37–44.
Muschet C, Möller G, Prehn C, et al. Removing the bottlenecks of cell culture metabolomics: fast normalization procedure, correlation of metabolites to cell number, and impact of the cell harvesting method. Metabolomics. 2016;12:151.
de Koning W, van Dam K. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal Biochem. 1992;204:118–23.
Villas-Bôas SG. Sampling and sample preparation. In: Villas-Bôas SG, Roessner U, Hansen MAE, et al, editors. Metabolome analysis: an introduction. Hoboken, NJ, USA: John Wiley & Sons, Inc. 2006. pp. 39–82.
Rizzi M, Baltes M, Theobald U, et al. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: II. Math Model Biotechnol Bioeng. 1997;55:592–608.
Schwiebert EM, Zsembery A. Extracellular ATP as a signaling molecule for epithelial cells. Biochim Biophys Acta Biomembr. 1615;2003:7–32.
Britten RJ, Mcclure FT. The amino acid pool in Escherichia coli. Bacteriol Rev. 1962;26:292.
Kostidis S, Addie RD, Morreau H, et al. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: a tutorial. Anal Chim Acta. 2017;980:1–24.
Madji Hounoum B, Blasco H, Nadal-Desbarats L, et al. Analytical methodology for metabolomics study of adherent mammalian cells using NMR, GC-MS and LC-HRMS. Anal Bioanal Chem. 2015;407:8861–72.
Matheus N, Hansen S, Rozet E, et al. An easy, convenient cell and tissue extraction protocol for nuclear magnetic resonance metabolomics. Phytochem Anal. 2014;25:342–9.
Zukunft S, Prehn C, Röhring C, et al. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics. 2018;14:1–12.
Beckonert O, Keun HC, Ebbels TMD, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–703.
Sellick CA, Hansen R, Stephens GM, et al. Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nat Protoc. 2011;6:1241–9.
Cao B, Aa J, Wang G, et al. GC–TOFMS analysis of metabolites in adherent MDCK cells and a novel strategy for identifying intracellular metabolic markers for use as cell amount indicators in data normalization. Anal Bioanal Chem. 2011;400:2983–93.
León Z, García-Cañaveras JC, Donato MT, et al. Mammalian cell metabolomics: experimental design and sample preparation. Electrophoresis. 2013;34:2762–75.
Merglen A, Theander S, Rubi B, et al. Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology. 2004;145:667–78.
Dingreville F, Panthu B, Thivolet C, et al. Differential effect of glucose on ER-mitochondria Ca2+ exchange participates in insulin secretion and glucotoxicity-mediated dysfunction of β-cells. Diabetes. 2019;68:1778–94.
Rieusset J, Fauconnier J, Paillard M, et al. Disruption of calcium transfer from ER to mitochondria links alterations of mitochondria-associated ER membrane integrity to hepatic insulin resistance. Diabetologia. 2016;59:614–23.
Akoka S, Barantin L, Trierweiler M. Concentration measurement by proton NMR using the ERETIC method. Anal Chem. 1999;71:2554–7.
Lu W, Su X, Klein MS, et al. Metabolite measurement: pitfalls to avoid and practices to follow. Annu Rev Biochem. 2017;86:277–304.
Sauerschnig C, Doppler M, Bueschl C, et al. Methanol generates numerous artifacts during sample extraction and storage of extracts in metabolomics research. Metabolites. 2017;8:1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1964 kb)
Rights and permissions
About this article
Cite this article
Mili, M., Panthu, B., Madec, AM. et al. Fast and ergonomic extraction of adherent mammalian cells for NMR-based metabolomics studies. Anal Bioanal Chem 412, 5453–5463 (2020). https://doi.org/10.1007/s00216-020-02764-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02764-9