Abstract
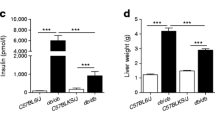
Insulin resistance (IR) lies at the origin of type 2 diabetes. It induces initial compensatory insulin secretion until insulin exhaustion and subsequent excessive levels of glucose (hyperglycemia). A high-calorie diet is a major risk factor contributing to the development of this metabolic disease. For this study, a time-course experiment was designed that consisted of two groups of mice. The aim of this design was to reproduce the dietary conditions that parallel the progress of IR over time. The first group was fed with a high-fatty-acid diet for several weeks and followed by 1 week of a low-fatty-acid intake, while the second group was fed with a low-fatty-acid diet during the entire experiment. The metabolomic fingerprint of C3HeB/FeJ mice liver tissue extracts was determined by means of two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC–ToF–MS). This article addresses the application of ANOVA-simultaneous component analysis (ASCA) to the found metabolomic profile. By performing hyphenated high-throughput analytical techniques together with multivariate chemometric methodology on metabolomic analysis, it enables us to investigate the sources of variability in the data related to each experimental factor of the study design (defined as time, diet and individual). The contribution of the diet factor in the dissimilarities between the samples appeared to be predominant over the time factor contribution. Nevertheless, there is a significant contribution of the time–diet interaction factor. Thus, evaluating the influences of the factors separately, as it is done in classical statistical methods, may lead to inaccurate interpretation of the data, preventing achievement of consistent biological conclusions.

Time-course design of the study. The experimental design recreates the typical IR progression over time sketched in the upper graph. Data from samples measured with GC×GC–ToF–MS were processed and ASCA analysis was performed to discern the different sources of variation in them. 1392 × 816 mm (150 × 150 DPI)





Similar content being viewed by others
References
Stumvoll M, Goldstein BJ, van Haeften TW (2005) Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365(9467):1333–1346. doi:10.1016/S0140-6736(05)61032-X
Marchesini G, Brizi M, Morselli-Labate AM, Bianchi G, Bugianesi E, McCullough AJ, Forlani G, Melchionda N (1999) Association of nonalcoholic fatty liver disease with insulin resistance. Am J Med 107(5):450–455. doi:10.1016/S0002-9343(99)00271-5
DeFronzo RA (2009) From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes 58(4):773–795. doi:10.2337/db09-9028
Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB (2009) Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes 58(11):2429–2443. doi:10.2337/db09-0580
Patti GJ, Yanes O, Siuzdak G (2012) Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 13(4):263–269
Xia J, Broadhurst D, Wilson M, Wishart D (2013) Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics 9(2):280–299. doi:10.1007/s11306-012-0482-9
Almstetter M, Oefner P, Dettmer K (2012) Comprehensive two-dimensional gas chromatography in metabolomics. Anal Bioanal Chem 402(6):1993–2013. doi:10.1007/s00216-011-5630-y
Wu Z, Huang Z, Lehmann R, Zhao C, Xu G (2009) The application of chromatography-mass spectrometry: methods to metabonomics. Chromatographia 69:23–32. doi:10.1365/s10337-009-0956-8
Koek M, van der Kloet F, Kleemann R, Kooistra T, Verheij E, Hankemeier T (2010) Semi-automated non-target processing in GC×GC–MS metabolomics analysis: applicability for biomedical studies. Metabolomics 7(1):1–14. doi:10.1007/s11306-010-0219-6
Zeng Z-D, Hugel H, Marriott P (2011) Chemometrics in comprehensive multidimensional separations. Anal Bioanal Chem 401(8):2373–2386. doi:10.1007/s00216-011-5139-4
Boccard J, Rudaz S (2014) Harnessing the complexity of metabolomic data with chemometrics. J Chemom 28(1):1–9. doi:10.1002/cem.2567
Smilde AK, Jansen JJ, Hoefsloot HCJ, Lamers R-JAN, van der Greef J, Timmerman ME (2005) ANOVA-simultaneous component analysis (ASCA): a new tool for analyzing designed metabolomics data. Bioinformatics 21(13):3043–3048. doi:10.1093/bioinformatics/bti476
Zwanenburg G, Hoefsloot HCJ, Westerhuis JA, Jansen JJ, Smilde AK (2011) ANOVA–principal component analysis and ANOVA–simultaneous component analysis: a comparison. J Chemom 25(10):561–567. doi:10.1002/cem.1400
Römisch-Margl W, Prehn C, Bogumil R, Röhring C, Suhre K, Adamski J (2011) Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics 8:1–10. doi:10.1007/s11306-011-0293-4
Birkemeyer C, Kolasa A, Kopka J (2003) Comprehensive chemical derivatization for gas chromatography-mass spectrometry-based multi-targeted profiling of the major phytohormones. J Chromatogr A 993(1–2):89–102. doi:10.1016/S0021-9673(03)00356-X
Grob K, Grob G, Grob K Jr (1981) Testing capillary gas chromatographic columns. J Chromatogr A 219(1):13–20
Bro R (2003) Multivariate calibration: what is in chemometrics for the analytical chemist? Anal Chim Acta 500(1–2):185–194. doi:10.1016/S0003-2670(03)00681-0
Wold S, Esbensen K, Geladi P (1987) Principal component analysis. Chemom Intell Lab Syst 2(1–3):37–52. doi:10.1016/0169-7439(87)80084-9
Trygg J, Holmes E, Lundstedt TR (2006) Chemometrics in metabonomics. J Proteome Res 6(2):469–479. doi:10.1021/pr060594q
van den Berg R, Hoefsloot H, Westerhuis J, Smilde A, van der Werf M (2006) Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7(1):142. doi:10.1186/1471-2164-7-142
Ester M, Kriegel H, Sander J, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. In: Simoudis E, Han J, Fayyad U (eds) Second International Conference on Knowledge Discovery and Data Mining. AAAI Press, pp 226–231. doi:citeulike-article-id:2265233
Jansen JJ, Hoefsloot HCJ, van der Greef J, Timmerman ME, Westerhuis JA, Smilde AK (2005) ASCA: analysis of multivariate data obtained from an experimental design. J Chemom 19(9):469–481. doi:10.1002/cem.952
Stehlík M, Střelec L, Thulin M (2014) On robust testing for normality in chemometrics. Chemom Intell Lab Syst 130(0):98–108. doi:10.1016/j.chemolab.2013.10.010
Cattell RB (1966) The scree test for the number of factors. Multivar Behav Res 1(2):245–276. doi:10.1207/s15327906mbr0102_10
Kahle M, Horsch M, Fridrich B, Seelig A, Schultheiß J, Leonhardt J, Irmler M, Beckers J, Rathkolb B, Wolf E, Franke N, Gailus-Durner V, Fuchs H, de Angelis MH, Neschen S (2013) Phenotypic comparison of common mouse strains developing high-fat diet-induced hepatosteatosis. Mol Metab 2(4):435–446. doi:10.1016/j.molmet.2013.07.009
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical collection Multidimensional Chromatography with guest editors Torsten C. Schmidt, Oliver J. Schmitz, and Thorsten Teutenberg.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1476 kb)
Rights and permissions
About this article
Cite this article
Ly-Verdú, S., Gröger, T.M., Arteaga-Salas, J.M. et al. Combining metabolomic non-targeted GC×GC–ToF–MS analysis and chemometric ASCA-based study of variances to assess dietary influence on type 2 diabetes development in a mouse model. Anal Bioanal Chem 407, 343–354 (2015). https://doi.org/10.1007/s00216-014-8227-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8227-4