Abstract
Over the last decade metabolomics has gained increasing popularity and significance in life sciences. Together with genomics, transcriptomics and proteomics, metabolomics provides additional information on specific reactions occurring in humans, allowing us to understand some of the metabolic pathways in pathological processes. Abnormal levels of such metabolites as nucleosides in the urine of cancer patients (abnormal in relation to the levels observed in healthy volunteers) seem to be an original potential diagnostic marker of carcinogenesis. However, the expectations regarding the diagnostic value of nucleosides may only be justified once an appropriate analytical procedure has been applied for their determination. The achievement of good specificity, sensitivity and reproducibility of the analysis depends on the right choice of the phases (e.g. sample pretreatment procedure), the analytical technique and the bioinformatic approach. Improving the techniques and methods applied implies greater interest in exploration of reliable diagnostic markers. This review covers the last 11 years of determination of urinary nucleosides conducted with the use of high-performance liquid chromatography in conjunction with various types of detection, sample pretreatment methods as well as bioinformatic data processing procedures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the most unpredictable diseases is cancer, especially if it is diagnosed in the advanced stage. Many diagnostic markers have been proposed but none have been proved so far to be sufficiently sensitive. That is why the mortality rate due to cancer is still increasing. For instance, carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 199, CA 125 and α-fetoprotein (AFP) were demonstrated to have insufficient sensitivity for testing for cancer and are currently used solely to detect a single type of the disease [1–3]. Moreover, the commonly available diagnostic tests, such as the test for prostate-specific antigen, can yield false-negative results.
Unlike the biochemical diagnostic approach, which focuses on single markers, metabolomics takes into account larger groups of metabolites and is therefore expected to limit the likelihood of false-negative results. In addition, it seems that a holistic approach (involving the analysis of metabolite profiles as a reflection of the individual’s health status) can provide a more reliable answer as to the presence or absence of cancer.
Nucleoside levels measured in urine samples may play a significant role as possible tumour markers. The justification of this hypothesis refers to a fundamental metabolic process, namely RNA turnover. In such pathological processes as blood disorders, inflammation, AIDS and malignancies, degradation of RNA is more rapid. The subsequent correlation between the presence of cancer and nucleoside levels might be due to the increased methyltransferase activity. As a result of both processes (increased RNA turnover and increased enzymatic activity), normal nucleosides undergo reutilisation and degradation to uric acid (as in the case of adenosine and guanosine), β-alanine (as in the case of cytidine and uridine) and β-aminoisobutyrate [4, 5]. Modified nucleosides, on the other hand, are not degraded or reutilised. Instead, they undergo distribution to the bloodstream and subsequent elimination with the urine in the unchanged form. Their abnormal urinary levels therefore correlate with either increased RNA turnover or increased methyltransferase activity, both of which are caused by cancer. This conclusion is not, however, clear and definitive. It cannot be presumed that patients with a more rapid RNA degradation have cancer or are at high risk of having cancer. They could be suffering from other disorders associated with increased cell turnover, such as chronic hepatitis and liver cirrhosis [6]. In terms of cancer these patients are initially considered healthy, but after some of the common tumour markers (e.g. AFP, CEA, CA 199 or CA 125) have been determined, they are reclassified as cancer patients. This may lead to false-positive results. One way of limiting such an undesirable situation is to use highly specific and sensitive biomarkers and to carefully select non-cancer volunteers. According to the literature, urinary nucleosides seem to be more specific and selective than the tumour markers commonly employed so far.
A large number of papers have emphasised that urinary nucleosides, especially modified ones, may be used as a diagnostic tool in various cancers, e.g. leukaemia [7], lymphoma [8], nasopharyngeal cancer, breast cancer [9–11], colorectal cancer [12, 13], bronchogenic carcinoma [14], bladder cancer [15–18] and hepatocellular carcinoma [6]. The fact that nucleoside levels do not depend on age and sex [5] suggests that these variables do not need to be taken into account when developing relevant diagnostic tests. On the other hand, nucleoside levels are also elevated in some physiological states, such as intensive body growth and pregnancy. All in all, it is important to emphasise that clinical use of nucleosides in the diagnosis of cancer is still confirmatory in nature and the results obtained from adequate analysis should be supported by the already available and widely used diagnostic methods, such as ultrasonography, cytology, histopathology and radiography.
The increased interest in metabolomics and the significant correlation between nucleosides and the processes implied by the presence of cancer have generated increasing interest in analytical and bioinformatic techniques involving these agents. Nucleosides can be analysed using various techniques according to their physicochemical properties, such as hydrophilic structure, molecular mass, electric charge over a wide pH range and ability to absorb both visible and UV light. The first paper on determination of urinary nucleosides, by Gehrke et al. [19], was published in 1978. The authors successfully determined six nucleosides in urine samples from healthy individuals and cancer patients using affinity liquid chromatography (LC) with a bound boronic acid stationary phase. Since that time, various techniques have been developed and applied to achieve greater sensitivity not only in chromatographic but also in electromigration methods. In this paper we review chromatographic determinations of the urinary nucleosides focusing particularly on papers published in the past 11 years.
Sample pretreatment procedure
Offline extraction
The sample pretreatment procedure is often the most important step in the entire analysis. The adaptation of an appropriate selective pretreatment method for analytes usually protects the matrix purification process from interferences.
To obtain well-purified and concentrated samples, the physicochemical properties and structures of the analytes need to be taken into account [20]. Numerous methods have been tested for satisfactory purification of nucleosides from the complex matrix of such biological samples as urine, serum, blood and even tissues. Nowadays, solid-phase extraction is often regarded as the best method. Boos et al. [21], for instance, proposed phenylboronic acid as an extraction agent for nucleosides because of its unique properties. This phase is selective for nucleosides thanks to the vicinal hydroxyl groups in their structure (during extraction the vicinal groups of nucleosides bind to the phenylboronic acid phase under basic conditions). While preserving basic conditions, one can purify samples from interferences without any loss of analytes. Only the alteration of pH from basic to acidic offers the chance to release analytes from the extraction phase and for them to be eluted afterwards. Thanks to that, the extracted and eluted analytes can be analysed further.
Many various modifications of phenylboronic acid as the extraction phase have been proposed over the years. Therefore, more and more reports are continuing to reveal that the phenylboronic acid phase is the method of choice for extraction of nucleosides. It should, however, be recognised that in the case of mixtures containing nucleosides without cis-diol groups, phenylboronic acid as the extraction phase does not play a significant role. In this case, other extraction phases, such as cation-exchange columns, are more suitable. Hsu et al. [22] proposed a cation-exchange MCX column with sulfate functional groups for nucleoside determination. This column is appropriate for all acidified nitrogen-containing nucleosides. Therefore, with this column it is possible to extract the deoxynucleosides which do not have vicinal groups in their structure, e.g. 2′-deoxyguanosine and 8-hydroxy-2′-deoxyguanosine [10, 22–26]. These two nucleosides are considered markers of oxidative DNA damage, which is also related to carcinogenesis.
Solid-phase extraction with C18 columns can also be applied for deoxynucleoside determination [7]. Moreover, according to a recent report [27], solid-phase microextraction seems to be a very promising method as far as nucleoside extraction is concerned. The method proposed by He et al. [27] relies on the high affinity of phenylboronic acid for these agents. The authors [27] used poly(3-aminophenylboronic acid) as the extraction material; this can selectively capture analytes with 1,2 or 1,3 cis-diol groups under basic conditions. A change of the pH from basic to acidic results in the previously captured analytes being subsequently released from the extraction phase and then eluted.
Online extraction
Sample pretreatment is usually considered a separate procedure to be carried out by the analyst. However, modern analytical techniques make it possible to automate it by integration within analytical equipment. The extraction is then performed online.
Hagemeier et al. [28] applied a method in which the standard extraction procedure was omitted and phenylboronic acid was substituted for the silica column in the process. The proposed analytical procedure was fully automated and based on using two coupled columns: a phenylboronic acid substituted silica column to clean and concentrate the analytes derived from the biological matrix, and a second column to perform the analytical chromatographic separation.
This approach has many advantages. For instance, the total analysis time is much shorter and the risk of specimen contamination or loss of analytes is limited. Following the example of Hagemeier et al. [28], more and more reports have suggested the use of a phenylboronic acid column directly connected to an analytical chromatographic one [2, 12, 29, 30].
The method developed by Cho et al. [25, 31] also did not require any separate pretreatment procedure. The authors efficiently removed interferences from the matrix using column-switching LC. Instead of a phenylboronic acid substituted silica column, they used an n-vinylacetamide copolymer column to clean the samples.
Instead of a phenylboronic acid column, Kowalska et al. [32] proposed a different column. They presented three different laboratory-made columns that may be useful for the analysis of nucleosides with competitive results.
Procedure without extraction
Although extraction seems to be the method of choice in the analysis of nucleosides, there are some applications that omit this step. Hewavitharana and Bruce [23] determined two analytes, creatinine and pseudouridine, without using any extraction methods before the chromatographic analysis. They analysed bovine blood treated with acetone and then mixed and centrifuged. To concentrate the analytes, the supernatant was evaporated and the residue was subsequently dissolved in an appropriate solvent directly before the analysis.
Another interesting approach has been proposed by Lee at al. [33], who developed an LC/electrospray ionisation (ESI) mass spectrometry (MS) method for urinary nucleosides, also without extraction. They showed that the analysis of nucleosides using the positive-ion mode with selected reaction monitoring effectively eliminated interferences from the endogenous constituents of the urine. This approach was both more convenient and more efficient in analysing large numbers of samples compared with the analytical methods involving complex extraction and/or derivatisation steps, as is the case with gas chromatography (GC) techniques [34].
Non-chromatographic techniques
Non-chromatographic methods are rarely used for the determination of nucleosides in biological samples. Some papers have reported on the use of immunoassays. Enzyme immunoassay used for the determination of normal and modified nucleosides is based on a specific complex reaction between the analyte and its unique antibody that is determined afterwards. This technique is considered specific and sensitive. However, it is not widely used because of limited applicability. So far it has only been performed for six nucleosides, for which specific antibodies have been developed: 5-methylcytidine, N 4-acetylcytidine 1-methylinosine, 1-methyladenosine, 7- methylguanosine and pseudouridine [8, 9, 35].
Chromatographic techniques
Nowadays, chromatographic techniques are well-established and commonly used in the analysis of nucleosides. Thanks to their structure, nucleosides can be determined by various types of detection, e.g. photometric, electrochemical and MS. In addition, according to the latest literature reports, the ultraperformance LC (UPLC) technique and the hydrophilic interaction chromatography (HILIC; also called “reverse reversed-phase” or “aqueous normal phase” chromatography) method with various modifications seem to be a good alternative for standard nucleoside analysis [24, 29].
The reported chromatographic methods for the determination of nucleosides are summarised in Tables 1 and S1 and a typical chromatogram of a nucleoside profile is presented in Fig. 1.
Chromatographic metabolite profiling analysis of a nucleoside standard mixture and b nucleosides extracted from urine. 1 pseudouridine, 2 cytidine, 3 uridine, 4 1-methyladenosine, 5 inosine, 6 5-methyluridine, 7 guanosine, 8 1-methylinosine, 9 1-methylguanosine, 10 N 4-acetylcytidine, 11 N 2-methylguanosine, 12 adenosine, 13 N 2,N 2-dimethylguanosine, 14 internal standard (8-bromoguanosine), 15 6-methyladenosine. (With permission from [12])
As presented in Tables 1 and S1, urinary nucleosides can be determined not only by high-performance LC (HPLC) and UPLC, but also by GC. However, in this case a very time consuming pretreatment procedure, which usually involves a hydrolytic reaction and/or a derivatisation step, renders this method rarely employed [34, 36].
Owing to their physiochemical properties, nucleosides are mainly analysed by reversed-phase HPLC with a C18 column. In this method, the mobile phase usually contains an appropriate buffer and an organic modifier: acetonitrile or methanol in various proportions (Table S2).
A good modern method of determination of nucleosides is UPLC. UPLC, which employs a column packed with 1.7-μm porous particles, seems to be more efficient than HPLC. The size of the particles provides a higher peak capacity, greater resolution and increased sensitivity. Zhao et al. [24] determined nucleosides by UPLC and HPLC with UV and MS detection. To compare HPLC and UPLC, the authors performed an analysis under identical conditions (Tables 1, S1, S2). As a result, 79 peaks were found in the case of HPLC-UV detection and 92 in case of UPLC-UV detection. UPLC yielded more peaks of high-performance efficiency. Regarding MS, it was possible to detect 11,000 ions (UPLC) and 8,000 ions (HPLC). The authors [24] also proved that the UPLC needed about one third of the time required by HPLC, while achieving a much better resolution and supplying more comprehensive information on metabolites.
Henriksen et al. [26] also used the UPLC-MS technique. They focused on the two nucleosides from urine samples, namely 8-oxoguanosine and 8-oxo-2′-deoxyguanosine, which play a pivotal role in oxidative DNA damage. The authors proposed a rapid (23 min) and automated method which can be useful for large-scale studies. The quantification limit in urine samples was 1 nM for both analytes. In addition, accuracy and precision were documented, showing average recoveries of 106.2% (8-oxoguanosine) and 106.9% (8-oxo-2′-deoxyguanosine), and the overall within-day and between-day precision was 6.1% and 4.4%, respectively.
The HILIC method is also worth considering. Its characteristic feature is the use of a polar stationary phase (e.g. silica, cyano) and a highly organic mobile phase (95–65% acetonitrile) with a smaller amount of aqueous solvent. In contrast to the composition of the mobile phase widely used in reversed-phase HPLC (Table S2), the aqueous solvent in HILIC is classified as a stronger eluent than the organic modifier. Therefore, this method can retain highly polar analytes that would not be retained by reversed-phase chromatography. Moreover, it shows a complementary selectivity for the reversed phase and shortens the sample preparation procedure: it makes it possible to omit the evaporation step following the usually performed extraction because the organic extract may be directly injected into the organic eluent.
Tuytten et al. [29] proposed a new approach using a HILIC column. They developed a method with an aprotic boronic acid precolumn, which was switched to the HILIC column: polyhydroxyethyl aspartamide. The mobile phase for the precolumn was 100% acetonitrile. Regarding the HILIC column, there were two eluents: eluent A was 10 mM ammonium formate in 95:4.8:0.2 acetonitrile/water/formic acid and eluent B was 10 mM ammonium formate in 50:49.8:0.2 acetonitrile/water/formic acid. The process was fully automated and allowed determination of three nucleosides in 40 min. Thanks to the aprotic boronic acid precolumn, the typical extraction procedure was eliminated and the analytes were well-purified and concentrated. However, this method can be adapted only for determination of nucleosides that are very soluble in acetonitrile, otherwise the high amount of acetonitrile in the initial mobile phase can cause precipitation of the analytes. It is also important to remember that every analyte determined with the use of the HILIC column should be dissolved in the solvent that is “weaker” than the initial mobile phase (or, in the worst case, in the initial mobile phase itself).
Although the HILIC method is designed for polar analytes that are not retained on the reversed-phase HPLC column, it is also appropriate for analytes soluble in high amounts of acetonitrile. That is why the physicochemical properties of the analytes and the sample origin (e.g. urine, blood) are pivotal factors taken into account in the selection of the HILIC method.
A different approach was proposed by Dudley et al. [36]. They compared three techniques: GC/MS, HPLC/ion trap MS (IT-MS) and capillary LC triple quadrupole MS (CapLC/TQMS) with either a full or a controlled neutral loss scan. All these methods proved to be valuable techniques for the analysis of urinary nucleosides.
In the case of GC/MS, the derivatisation step was required, which significantly prolonged the total analysis time. However, thanks to the derivatisation step, dihydrouridine and 3-methylcytidine could be determined, whereas in the case of the LC techniques neither was detected. Moreover, each method detected two singly methylated guanosines and one dimethylguanosine, whereas the two LC/MS methods detected another analyte, namely a trimethylated guanosine. Similarly to GC/MS, CapLC/TQMS was also very time consuming because of the long reequilibration time (1 h) in comparison with HPLC (15 min).
Dudley et al. [36] compared the sensitivity of the methods, and GS/MS was shown to be tenfold less sensitive than HPLC/IT-MS in the analysis of nucleosides. CapLC/TQMS with the full scan mode, in turn, offered a vast improvement in sensitivity. The sensitivity was still better than that of HPLC/IT-MS when constant neutral loss mode of scanning was applied, although the difference in sensitivity between full and constant neutral loss modes was very significant. It is important to note that the authors proposed applying each method instead of choosing only one because the methods used together provide valuable information.
Type of detection
Up to now, most of the methods have been based on UV spectrophotometric detection with a wavelength usually set at 254 nm [1, 2, 7, 10, 12, 13, 23, 37–43]. However, this detection method is not completely satisfactory as, for instance, dihydrouridine absorbs light at 210 nm [44]. To overcome this problem a diode-array detector, which allows measurements over the full range of the UV spectrum, can be applied.
An interesting approach was proposed by Kelly et al. [45]. They determined products of oxidative stress using HPLC coupled with both UV and electrochemical detection. Electrochemical detection allows for a specific determination of oxidation products, 8-oxoguanosine and 8-hydroxydeoxyguanosine, which is not possible with simple UV detection. However, UV detection allows for simultaneous detection of unmodified products. The advantages of the proposed method were the short time of analysis and good resolution. Hence, it can be used as an alternative approach for determination of urinary nucleosides, especially those which reflect oxidative DNA damage.
It is worth noting that spectrophotometric detection is not very sensitive, especially when nucleosides are determined in biological samples. It is also well known that to confirm the identity of each nucleoside reference standards are required. Unfortunately, only several of them are commercially available. Hence, this type of detection is not satisfactory because of both poor selectivity and poor detection limits. To improve the sensitivity and determine all the desired nucleosides in urine samples, MS detection should be employed.
HPLC/MS analysis has several advantages over the previously discussed detection methods. The coupling of HPLC instrumentation to a mass spectrometer facilitates further identification of the nucleoside by identifying the nucleoside’s quasi-molecular ion, thereby allowing mass determination, and by MS/MS fragmentation, providing structural information about the nucleoside. HPLC/MS also allows quantitation of more than one nucleoside at a time, unlike some of the methods applied earlier, such as immunoassay [35].
As far as determination of urinary nucleosides is concerned, MS is very often used with great success. In most reports, ESI in positive mode has been described [22, 24–26, 31, 33, 34, 46]. Dudley et al. [47] checked both negative and positive ionisation modes. The intensity was far greater in the positive than in the negative ionisation mode.
Bullinger et al. [48] focused on a different approach from those described in the other reports. The isolated metabolites were analysed by IT-MS and Fourier transform ion cyclotron resonance MS. As a result, the authors identified 22 metabolites deriving from cellular RNA metabolism and related metabolic pathways, among which four modified nucleosides were found for the first time, namely 2-methylthio-N 6-(cis-hydroxyisopentenyl)-adenosine, 5-methoxycarbonylmethyl-2-thiouridine, N 6-methyl-N 6-threonylcarbamoyladenosine and 2-methylthio-N 6-threonylcarbamoyladenosine.
Kammerer et al. [49] developed an auto-LC/MS3 method. Whereas the MS/MS spectra only show the nucleic base fragment, the MS3 fragmentation step provides further, valuable information about the structure of the nucleic base itself. Therefore, the authors were able to identify 15 nucleosides by their retention times and three by comparing the fragmentation with that of standard substances.
Kammerer et al. [42] used matrix-assisted laser desorption/ionisation (MALDI) time-of-flight MS with a 337-nm nitrogen laser to identify nucleosides. As the matrix substances, 2,5-dihydroxybenzoic acid and α-cyano-4-hydroxycinamic acid (CHCA) were used. Postsource decay (PSD) spectra of the molecular ions were also recorded. The PSD spectra of the nucleosides showed a characteristic cleavage between the sugar moiety and the nucleic base. Depending on the basicity of the nucleoside, using CHCA as the matrix, the detection limit ranged from 100 fmol to 10 pmol.
Apart from good sensitivity, there is also a disadvantage of using this method. MALDI is an offline technique with increased sample preparation requirements: sample preparation for MALDI analysis is quite a time-consuming procedure.
Chemometric processing of nucleoside separation data
The large data sets obtained after each analysis contain not only chemical results but also inexplicit information about patients, for instance, sex, age, health and disease status. Therefore, it appears advisable to take into account every kind of data available, whether straightforward or hidden in multidimensional data sets.
To correlate analytical data with the physiological or pathophysiological status of the donor of the analysed biological material, a simple mathematical procedure cannot suffice. However, thanks to the advanced bioinformatic methods which can cope with vast data sets, it is possible to extract systematic information originally dispersed over extensive data matrices. Over the last decade, the bioinformatic approach has been improved and some previously unresolved technical problems are nowadays simple and easy to handle.
In regard to urinary nucleosides, the most important challenge is the confirmation that nucleosides can serve as universal markers in urogenital cancer. In some forms of cancer it is speculated that nucleosides are potential disease markers. For instance, pseudouridine is a far-sighted factor in colorectal cancer. Other nucleosides, such as 1-methyladenosine, 5-hydroxymethyl-2′-deoxyuridine and 8-hydroxy-2-deoxyguanosine, may play the role of biomarkers in breast cancer patients.
Zheng et al. [2] verified the hypothesis of significant differences in the levels of nucleosides in various stages of cancer. They [2] determined 14 nucleosides derived from urine samples obtained from (1) colorectal cancer patients, (2) patients with intestinal villous adenoma and (3) healthy adults. When the mean values for the three groups were analysed, the most abundant nucleoside appeared to be pseudouridine, followed by 1-methyladenosine and 1-methylinosine. The pseudouridine level was elevated above the normal values plus two standard deviations in 58% (30/52) of patients with colorectal cancer, whereas in 20% (2/10) of patients with intestinal villous adenoma it was just elevated. It was also found that the level of these nucleosides positively correlated with the tumour size and Duke’s stage of colorectal cancer (p < 0.05). However, no significant correlations were noticed for the observed values with regard to tumour differentiation. The concentrations of 12 nucleosides (except uridine and guanosine) were significantly elevated in patients with colorectal cancer (p < 0.05) (Tables 2, 3).
The second aim of the study by Zheng et al. [2] was to verify whether there were significant changes in the concentrations of nucleosides before and after surgery. The patients were classified into two groups: the response group (32 patients) and the progressive disease group (11 patients). Using the paired t test, the concentrations of such nucleosides as pseudouridine, 1-methyladenosine, 1-methylinosine, 1-methylguanosine, N-4-acetylocytidine and dimethylguanosine from the response group before surgery were significantly higher than those of patients after surgery (p < 0.05). However, the presurgery and postsurgery difference in the levels of these nucleosides for the progressive disease group showed no significant changes.
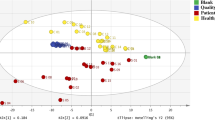
Zheng et al. [2] also compared the sensitivity of urinary nucleosides in patients with colorectal cancer with the sensitivity of the commonly used tumour markers, such as CEA. Using concentrations of 14 nucleosides as the data vectors for principal component analysis (PCA), they distinguished 71% (37/52) of the patients with colorectal cancer from healthy adults, whereas healthy adults were correctly classified at 96% (58/60) specificity (Fig. 2). Taking into account the specificity of the CEA method, which was 29% (15/52), it seems highly probable that nucleosides may play an important role as a predictive tool in carcinogenesis.
Principal component analysis of urine nucleoside profiles from colorectal cancer patients (circles) and healthy controls (crosses). Asterisks a positions of patients with intestinal villous adenoma, b positions of 32 responsive cases after surgery, c positions of 11 progressive cases after surgery. PC principal component. (With permission from [2])
In parallel with the above report, Feng et al. [1] confirmed a significant biomarker role of urinary nucleosides in patients with colorectal cancer. When the PCA method was used, higher levels of 11 out of 14 nucleosides were demonstrated in cancer patients compared with healthy individuals. In addition to the concentration of 1-methyladenosine, the concentrations of pseudouridine and 1-methylguanosine were much higher in patients with colorectal cancer and may also be promising as diagnostic tools soon (Table 2, 3). The results were compared with the commonly used tumour markers, such as CEA, CA 199, CA 125 and AFP. The sensitivity of the analysis was much higher (76.9%) than that of CEA (38.5%), CA 199 (40.4%), CA 125 (15.4%) and AFP (17.3%). These results confirm that the value of urinary nucleosides has already been appreciated.
The first report on using HPLC/ESI-MS/MS in the analysis of nucleosides in urine samples from patients with colorectal cancer was by Hsu et al. [22]. They determined nine nucleosides (cytidine, 3-methylcytidine, 1-methyladenosine, uridine, adenosine, inosine, 2-deoxyguanosine, 8-hydroxy-2′-deoxyguanosine and N,N-dimethylguanine) in urine samples from cancer patients and healthy volunteers. The selected reaction monitoring mode was used in this study and therefore made it more specific and sensitive for quantitative analysis (Fig. 3). To increase the reliability of the data, each corrected mean concentration of the nucleosides in the cancer group was converted to a percentage of the corresponding normal mean value. In the reviewed study, the t test and Pearson’s test were used to measure the differences between cancer patients and the healthy controls (p < 0.05). It was showed that five urinary nucleosides may play a pivotal role in colorectal cancer, namely cytidine, adenosine, 8-hydroxy-2′-deoxyguanosine, N,N-dimethylguanine and uridine. It was also confirmed that a higher level of 8-hydroxy-2′-deoxyguanosine could be correlated with oxidative DNA damage, which is also considered to be a consequence of carcinogenesis.
Mass chromatograms of nine nucleosides and internal standard under selected reaction monitoring operation. ISTD internal standard. (With permission from [22])
In another report, the level of 8-hydroxy-2′-deoxyguanosine was decreased in cancer patients undergoing surgery and chemotherapy compared with the baseline [50]. Hence, the search for the possible tumour markers should not only focus on nucleoside metabolites but also on deoxynucleosides.
With regard to other cancers, Xu et al. [39] tested if urinary nucleosides could be used for the diagnosis of breast cancer. To estimate the origin of the sample, which may be derived from cancer patients or from healthy controls, a factor analysis was proposed. It was demonstrated that the levels of pseudouridine, 1-methyladenosine, 1-methylinosine and 1-methylguanosine were significantly higher, which should be taken into account in future research. Moreover, the level of 5-methyluridine was also much higher in cancer patients than in healthy volunteers, but the small normal sample did not allow certainty of the significance of that correlation.
Cho et al. [25] went further and determined the urinary levels of nucleosides in breast cancer patients before and after tumour removal. They measured urinary nucleosides from three groups: (1) healthy individuals, (2) breast cancer patients awaiting surgery and (3) the same patients following tumour removal. Pre- and postoperative samples were collected just before and 2 weeks after surgery and were directly analysed by column-switching LC/MS/MS (Fig. 4). The data were statistically assessed using the Mann–Whitney U test (p < 0.05). It was shown that the levels of nucleosides, such as 1-methyladenosine (p < 0.02), N 2,N 2-dimethylguanosine (p < 0.001), 5-hydroxymethyl-2′-deoxyuridine (p < 0.001) and 8-hydroxy-2′-deoxyguanosine (p < 0.001), were significantly elevated in preoperative patients compared with normal controls. The levels of the four modified nucleosides were lower in postsurgery versus presurgery patients (1-methyladenosine, N 2,N 2-dimethylguanosine, 5-hydroxymethyl-2′-deoxyuridine and 8-hydroxy-2′-deoxyguanosine). Furthermore, decreased concentrations of the four modified nucleosides after surgery were observed. In a similar comparison study, postsurgery concentrations were still higher than in normal controls. These results are consistent with the report by Xu et al. [10], in which the same three out of four nucleosides discussed by Cho et al. [25] were reported as showing elevated levels (1-methyladenosine, 5-hydroxymethyl-2′-deoxyuridine and 8-hydroxy-2-deoxyguanosine).
liquid chromatography/tandem mass spectrometry chromatograms for urinary nucleosides from breast cancer patients before (dotted line) and after (solid line) surgery. RT retention time. (With permission from [25])
Bullinger et al. [37] proposed different methods. They used the k nearest neighbour and support vector machine (SVM) methods within the leave-one-out procedure to predict the most significant nucleosides in breast cancer patients. They found elevated concentrations of 1-methylguanosine, 1-methylinosine, inosine, 3-methyluridine and adenosine. According to the Battiti–Tesmer algorithm, such nucleosides as pseudouridine, uridine, inosine, 3-methyluridine were selected, which suggested some inner redundancy in the measured nucleoside profiles. A large number (over 200) of biological samples were assayed and the results were statistically processed by Frickenschmidt et al. [46]. The 113 urinary samples derived from patients with breast cancer and 99 control samples from healthy volunteers were studied using LC/ESI-IT-MS. From each sample, 31 compounds were statistically analysed using PCA and SVM. PCA seemed to be inappropriate for such a large data set (212 samples × 31 compounds) because the resulting classification of the samples was unclear and chaotic. There were no clusters that should have represented cancer patients and healthy volunteers, which is usually observed in the analysis of smaller numbers of samples (fewer than 100). Hence, Frickenschmidt et al. [46] proposed another statistical model based on SVM with a recursive feature elimination algorithm. The sensitivity and specificity of the results were 89.39% and 88.89%, respectively, which is a significant improvement compared with the widely used breast cancer markers, such as CA 15–3 (specificity 90–95%, sensitivity 50%). Moreover, it should be taken into account that both the specificity and the selectivity of the nucleosides are the most important factors as regards their role in the diagnosis of cancer. However, the second equally important aspect is the number of data in the set on which the analysis is performed: the higher the analysed number of samples, the more statistically significant the results and the more credible the role of nucleosides as cancer markers.
Dieterle et al. [40] classified samples from healthy individuals and breast cancer patients using two different implementations of learning vector quantification (LVQ) and dynamic LVQ . They also adapted SVM backpropagations. These three methods were compared in terms of their sensitivity, specificity and prediction rate. Clinical sensitivity was calculated as the percentage of true negatives and the prediction rate was the percentage of true positives and true negatives in all individuals. As a result, LVQ classified 60 out of 85 cancer patients and 109 out of 121 healthy patients correctly. The sensitivity, specificity and prediction rate were approximated at 70.6%, 90.1% and 82.1%, respectively. The prediction rate of backpropagation was practically identical to that of LVQ, with exactly the same specificity and a slightly higher sensitivity. The prediction rate of SVM was also very similar to the prediction rate of LVQ, with slightly lower sensitivity and selectivity. The Wilcoxon test, also applied in this study, showed no significant differences between the methods. However, it was recommended to use LVQ for a further, more extended study.
The artificial neural network (ANN) technique seems to be a very useful tool in the diagnosis of cancer. In their study, Seidel et al. [13] used a multilayer perceptron (MLP) artificial neural network and non-parametric statistical tests, namely the Kolmogorov–Smirnov test along with the two-tailed Mann–Whitney test. The samples were collected from patients suffering from different cancers. The study groups consisted of 55 patients with histologically documented malignancies (26 patients with breast cancer, eight with colorectal cancer, seven with sarcoma, three with thyroid cancer, three with Hodgkin’s disease, two with melanoma, two with bronchial carcinoma, two with gynaecological cancers and two with other malignancies), 13 patients with benign diseases (nine with thyroid adenoma, two with chondromas, one with lipoma and one with parathyroid adenoma) and 41 healthy controls. To compare all the cancer samples with the healthy ones, Seidel et al. [13] used four input neurons and subsequently achieved a specificity of 85.4% and a sensitivity of 97%. Moreover, they distinguished benign from malignant tumours by MLP with also four input neurons and then achieved a specificity and sensitivity of 67% and 86%, respectively. The ANN technique was also used by Yang et al. [43]. They analysed urine samples (n = 69) and serum samples (n = 42) from cancer patients and healthy controls. Each sample (defined as a pattern) was described by 15 feature variables, which were the concentrations of 15 nucleosides. For the training set, an algorithm called “gradient descent with momentum and adaptive learning rate backpropagation” was used. The sensitivity and specificity calculated for the urine samples were 98% and 94.7%, respectively, and those for the serum samples were 94.7% and 95.8%, respectively. The authors also used the PCA technique to compare the results with ANN. The PCA technique appeared to be less sensitive and specific than ANN. For the urine data set, the sensitivity and the specificity were 92.2% and 77.8%, respectively, whereas those for the serum were 73.7% and 79.2%, respectively.
Summary and conclusions
The ongoing development of both analytical techniques and bioinformatic methods is generating increasing interest in metabolomics. Nowadays, the application of analytical techniques is heading towards the reduction of the total analysis time and implementation of fully automated systems with the most sensitive analytical detection methods that enable quantification at a very low concentration of analytes. It is very important to conduct such research, as so far many peaks obtained in the separation of biological samples have still not been identified. Certainly the use of a sensitive and selective method can solve that problem. Nevertheless, the development level of analytical procedures so far, although giving complex analytical data, is still not high enough to estimate the biological value of nucleosides as cancer markers. Only the use of sensitive and modern analytical techniques in connection with bioinformatic methods can facilitate the interpretation of complex analytical data and therefore make it possible to highlight the significant role of nucleosides as cancer biomarkers. To prove the role of nucleosides in carcinogenesis, many samples should be determined at an acceptably low concentration and then statistically evaluated. Basically, the large data sets obtained from either healthy volunteers or cancer patients are gathered in databases and can be analysed further by statistical and chemometric methods. Such databases consist of a variety of information (sex, age, additional dysfunctions, disease stage along with the analytical results). Their interpretation without bioinformatic tools can only be superficial. Eventually, analytical procedures combined with bioinformatic methods may answer the question about the actual diagnostic relevance of nucleosides. Given the results obtained so far, it seems that sooner or later this question will have a definite answer. To make this a reality, large numbers of biological samples should be determined and statistically processed.
References
Feng B, Zheng MH, Zheng YF, Lu AG, Li JW, Wang ML, Ma JJ, Xu GW, Liu BY, Zhu ZH (2005) J Gastrol Hepatol 20:1913–1919
Zheng YF, Yang J, Zhao XJ, Feng B, Kong HW, Chen YJ, Lv S, Zheng MH, Xu GW (2005) World J Gastroenterol 11:3871–3876
Liebich HM, Lehmann R, Xu G, Wahl HG, Häring HU (2000) J Chromatogr B 745:189–196
Limbach PA, Crain PF, McCloskey JA (1994) Nucleic Acids Res 22:2183–2196
Liebich HM, Müller-Hagedorn S, Bacher M, Scheel–Walter HG, Lu X, Frickenschmidt A, Kammerer B, Kim KR, Gérard H (2005) J Chromatogr B 814:275–283
Yang J, Xu G, Zheng Y, Kong H, Pang T, Lv S, Yang Q (2004) J Chromatogr B 813:59–65
Zambonin CG, Aresta A, Palmisano F, Specchia G, Liso V (1999) J Pharm Biomed Anal 21:1045–1051
Rasmunson T, Björk GR (1995) Acta Oncol 34:61–67
Sasco AJ, Rey F, Reynaud C, Bobin YJ, Clavel M, Niveleau A (1996) Cancer Lett 108:157–162
Xu G, Di Stefano C, Liebich HM, Zhang Y, Lu P (1999) J Chromatogr B 732:307–313
Zheng YF, Kong HW, Xiong JH, Lv S, Xu GW (2005) Clin Biochem 38:24–30
Zheng Y, Xu G, Yang J, Zhao X, Pang T, Kong H (2005) J Chromatogr B 819:85–90
Seidel P, Seidel A, Herbarth O (2007) Neural Netw 20:646–651
Waalkes TP, Abeloff MD, Ettinger DS, Woo KB, Gehrke CW, Kuo KC, Borek E (1982) Eur J Cancer Clin Oncol 18:1267–1274
Xie G, Su M, Li P, Gu X, Yan C, Qiu Y, Li H, Jia W (2007) Electrophoresis 28:4459–4468
Wang S, Zhao X, Mao Y, ChengY (2007) J Chromatogr A 1147:254–260
Szymańska E, Markuszewski MJ, Capron X, van Nederkassel AM, Vander Heyden Y, Markuszewski M, Krajka K, Kaliszan R (2007) Electrophoresis 43:413–420
Szymańska E, Markuszewski MJ, Bodzioch K, Kaliszan R (2007) J Pharm Biomed Anal 44:1118–1126
Gehrke CW, Kuo KC, Davis GE, Suits RD, Waalkes TP, Borek E (1978) J Chromatogr 150:455–476
Tuytten R, Lemière F, Esmans EL, Herrebout WA, van der Veken BJ, Maes BU, Witters E, Newton RP, Dudley E (2007) Anal Chem 79:6662–6669
Boss KS, Wilmers B, Schlimme E, Sauerbrey R (1988) J Chromatogr 456:93–104
Hsu WY, Chen WTL, Lin WD, Tsai FJ, Tsai Y, Lin CT, Lo WY, Jeng LB, Lai CC (2009) Clin Chim Acta 402:31–37
Hewavitharana AK, Bruce HL (2003) J Chromatogr B 784:275–281
Zhao X, Wang W, Wang J, Yang J, Xu G (2006) J Sep Sci 29:2444–2451
Cho SH, Choi MH, Lee WY, Chung BC (2009) Clin Biochem 42:540–543
Henriksen T, Hillestrøm PR, Poulsen HE, Weimann A (2009) FRBM 47:629–635
He J, Liu Z, Dou P, Liu J, Ren L, Chen HY (2009) Talanta 79:746–751
Hagemeier E, Boos KS, Schlimme E, Lechtenborger K, Kettrup A (1983) J Chromatogr 268:291–295
Tuytten R, Lemière F, Van Dongen W, Witters E, Esmans EL, Newton RP, Dudley E (2008) Anal Chem 80:1263–1271
Li F, Zhao X, Wang W, Xu G (2006) Anal Chim Acta 580:181–187
Cho SH, Jung BH, Lee SH, Lee WY, Kong G, Chung BC (2006) Biomed Chromatogr 20:1229–1236
Kowalska S, Krupczyńska K, Buszewski B (2005) J Sep Sci 28: 1502–1511
Lee SH, Jung BH, Kim SY, Chung BC (2004) Rapid Commun Mass Spectrom 18:973–977
Woo HM, Kim KM, Choi MH, Jung BH, Lee J, Kong G, Nam SJ, Kim S, Bai SW, Chung BC (2009) Clin Chim Acta 400:63–69
Honda I, Itoh K, Mizugaki M, Sasaki T (1999) Tohoku J Exp Med 188:133–138
Dudley E, Lemiere F, Van Dongen W, Langridge JI, El-Sharkawi S, Games DE, Esmans EL, Newton RP (2001) Rapid Commun Mass Spectrom 15:1701–1707
Bullinger D, Fröchlich H, Klaus F, Neubauer H, Frickenschmidt A, Henneges C, Zell A, Laufer S, Gleiter CH, Liebich H, Kammerer B (2008) Anal Chim Acta 618:29–34
Liebich HM, Müller-Hagedorn S, Klaus F, Meziane K, Kim KR, Frickenschmidt A, Kammerer B (2005) J Chromatogr A 1071:271–275
Xu G, Schmid HR, Lu X, Liebich HM, Lu P (2000) Biomed Chromatogr 14:459–463
Dieterle F, Müller-Hagedorn S, Liebich HM, Gauglitz G (2003) Art Intell Med 28:265–279
Xu G, Enderle H, Liebich H, Lu P (2000) Chromatographia 52:152–158
Kammerer B, Frickenschmidt A, Gleiter CH, Laufer S, Liebich H (2005) J Am Soc Mass Spectrom 16:940–947
Yang J, Xu G, Kong H, Zheng Y, Pang T, Yang Q (2002) J Chromatogr B 780:27–33
Xing F, Hiley SL, Hughes TR, Phizicky EM (2004) J Biol Chem 279:17850–17860
Kelly MC, White B, Smyth MR (2008) J Chromatogr B 863:181–186
Frickenschmidt A, Fröhlich H, Bullinger D, Zell A, Laufer S, Gleiter CH, Liebich H, Kammerer B (2008) Biomarkers 13:435–449
Dudley E, El-Sharkawi S, Games DE, Newton RP (2000) Rapid Commun Mass Spectrom 14:1200–1207
Bullinger D, Fux R, Nicholson G, Plontke S, Belka C, Laufer S, Gleiter CH, Kammerer B (2008) J Am Soc Mass Spectrom 19:1500–1513
Kammerer B, Frickenschmidt A, Müller CE, Laufer S, Gleiter CH, Liebich H (2005) Anal Bioanal Chem 382:1017–1026
Mei S, Yao Q, Wu C, Xu G (2005) J Chromatogr B 827:83–87
Acknowledgement
The project was supported by the Ministry of Science and Higher Education, Warsaw, Poland (grant numbers N405 101334 and N405 630338).
Conflict of interest
The authors declare no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the special issue Biomarkers with Guest Editors Boguslaw Buszewski and Jochen Schubert.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 113 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Struck, W., Waszczuk-Jankowska, M., Kaliszan, R. et al. The state-of-the-art determination of urinary nucleosides using chromatographic techniques “hyphenated” with advanced bioinformatic methods. Anal Bioanal Chem 401, 2039–2050 (2011). https://doi.org/10.1007/s00216-011-4789-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-011-4789-6