Abstract
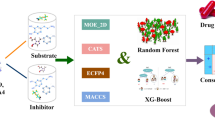
Cytochrome P450 enzymes are a superfamily of enzymes responsible for the metabolism of a variety of medicines and xenobiotics. Among the Cytochrome P450 family, five isozymes that include 1A2, 2C9, 2C19, 2D6, and 3A4 are most important for the metabolism of xenobiotics. Inhibition of any of these five CYP isozymes causes drug-drug interactions with high pharmacological and toxicological effects. So, the inhibition or non-inhibition prediction of these isozymes is of great importance. Many techniques based on machine learning and deep learning algorithms are currently being used to predict whether these isozymes will be inhibited or not. In this study, three different molecular or substructural properties that include Morgan, MACCS and Morgan (combined) and RDKit of the various molecules are used to train a distinct SVM model against each isozyme (1A2, 2C9, 2C19, 2D6, and 3A4). On the independent dataset, Morgan fingerprints provided the best results, while MACCS and Morgan (combined) achieved comparable results in terms of balanced accuracy (BA), sensitivity (Sn), and Mathews correlation coefficient (MCC). For the Morgan fingerprints, balanced accuracies (BA), Mathews correlation coefficients (MCC), and sensitivities (Sn) against each CYPs isozyme, 1A2, 2C9, 2C19, 2D6, and 3A4 on an independent dataset ranged between 0.81 and 0.85, 0.61 and 0.70, 0.72 and 0.83, respectively. Similarly, on the independent dataset, MACCS and Morgan (combined) fingerprints achieved competitive results in terms of balanced accuracies (BA), Mathews correlation coefficients (MCC), and sensitivities (Sn) against each CYPs isozyme, 1A2, 2C9, 2C19, 2D6, and 3A4, which ranged between 0.79 and 0.85, 0.59 and 0.69, 0.69 and 0.82, respectively.







Similar content being viewed by others
References
Abbas Z, Rehman MU, Tayara H, Chong KT (2023a) ORI-explorer: a unified cell-specific tool for origin of replication sites prediction by feature fusion. Bioinformatics 39(11):btad664
Abbas Z, ur Rehman M, Tayara H, Zou Q, Chong KT (2023b) XGBoost framework with feature selection for the prediction of RNA N5-methylcytosine sites. Mol Ther 31:2543–2551
Ahmad W, Tayara H, Chong KT (2023) Attention-based graph neural network for molecular solubility prediction. ACS Omega 8(3):3236–3244
Ahmad W, Tayara H, Shim H, Chong KT (2024) SolPredictor: predicting solubility with residual gated graph neural network. Int J Mol Sci 25(2):715
Ai D, Cai H, Wei J, Zhao D, Chen Y, Wang L (2023) DEEPCYPs: a deep learning platform for enhanced cytochrome P450 activity prediction. Front Pharmacol 14:1099093
Amarappa S, Sathyanarayana S (2014) Data classification using support vector machine (SVM), a simplified approach. Int J Electron Comput Sci Eng 3:435–445
Arimoto R (2006) Computational models for predicting interactions with cytochrome P450 enzyme. Curr Top Med Chem 6(15):1609–1618
Banerjee P, Dehnbostel FO, Preissner R (2018) Prediction is a balancing act: importance of sampling methods to balance sensitivity and specificity of predictive models based on imbalanced chemical data sets. Front Chem 6:387941
Banerjee P, Dunkel M, Kemmler E, Preissner R (2020) SuperCYPspred—a web server for the prediction of cytochrome activity. Nucleic Acids Res 48(W1):W580–W585
Bhagat M, Bakariya B (2022) Implementation of logistic regression on diabetic dataset using train-test-split, k-fold and stratified k-fold approach. Natl Acad Sci Lett 45(5):401–404
Boughorbel S, Jarray F, El-Anbari M (2017) Optimal classifier for imbalanced data using Matthews correlation coefficient metric. PLoS ONE 12(6):e0177678
Chen Y, Zeng L, Wang Y, Tolleson WH, Knox B, Chen S, Ren Z, Guo L, Mei N, Qian F, Huang K, Liu D, Tong W, Yu D, Ning B (2017) The expression, induction and pharmacological activity of CYP1A2 are post-transcriptionally regulated by microRNA hsa-miR-132-5p. Biochem Pharmacol 145:178–191
Cheng F, Yu Y, Shen J, Yang L, Li W, Liu G, Lee PW, Tang Y (2011) Classification of cytochrome P450 inhibitors and noninhibitors using combined classifiers. J Chem Inf Model 51(5):996–1011
Corsini A, Bortolini M (2013) Drug-induced liver injury: the role of drug metabolism and transport. J Clin Pharmacol 53(5):463–474
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Gaffar S, Hassan MT, Tayara H, Chong KT (2024) IF-AIP: a machine learning method for the identification of anti-inflammatory peptides using multi-feature fusion strategy. Comput Biol Med 168:107724
Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrián-Uhalte E et al (2017) The ChEMBL database in 2017. Nucleic Acids Res 45(D1):D945–D954
Guengerich FP (2008) Cytochrome P450 and chemical toxicology. Chem Res Toxicol 21(1):70–83
Hassan MT, Tayara H, Chong KT (2023) Meta-IL4: an ensemble learning approach for IL-4-inducing peptide prediction. Methods 217:49–56
Hassan MT, Tayara H, Chong KT (2024) An integrative machine learning model for the identification of tumor T-cell antigens. BioSystems 237:105177
Kato H (2020) Computational prediction of cytochrome P450 inhibition and induction. Drug Metab Pharmacokinet 35(1):30–44
Lakshmanan M (2019) Drug metabolism. In: Raj GM, Raveendran R (eds) Introduction to basics of pharmacology and toxicology: volume 1: general and molecular pharmacology: principles of drug action. Springer, Singapore, 99–116
Landrum G (2013) RDKit documentation. Release 1(1–79):4
Lazarou J, Pomeranz BH, Corey PN (1998) Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 279(15):1200–1205
Li X, Xu Y, Lai L, Pei J (2018) Prediction of human cytochrome P450 inhibition using a multitask deep autoencoder neural network. Mol Pharm 15(10):4336–4345
Lynch T, Price A (2007) The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Phys 76(3):391–396
Mahesh T, Geman O, Margala M, Guduri M et al (2023) The stratified K-folds cross-validation and class-balancing methods with high-performance ensemble classifiers for breast cancer classification. Healthc Anal 4:100247
National Center for Advancing Translational Sciences (NCATS) (2007) Bioassay record: P450-CYP1A2. PubChem. Accessed 5 Jan 2024
National Center for Advancing Translational Sciences (NCATS) (2009) Cytochrome panel assay with activity outcomes. PubChem BioAssay AID 1851. Accessed 5 Jan 2024
National Center for Advancing Translational Sciences (NCATS) (2010a) qHTS assay for inhibitors and substrates of cytochrome P450 2C19. PubChem. Accessed 5 Jan 2024
National Center for Advancing Translational Sciences (NCATS) (2010b) qHTS assay for inhibitors and substrates of cytochrome P450 2C9. PubChem. Accessed 5 Jan 2024
National Center for Advancing Translational Sciences (NCATS) (2010c) qHTS assay for inhibitors and substrates of cytochrome P450 2D6. PubChem. Accessed 5 Jan 2024
National Center for Advancing Translational Sciences (NCATS) (2010d) qHTS assay for inhibitors and substrates of cytochrome P450 3A4. PubChem. Accessed 5 Jan 2024
Ogu CC, Maxa JL (2000) Drug interactions due to cytochrome P450. In: Proceedings (Baylor University. Medical Center), vol 13 no. 4. p 421
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V et al (2011) Scikit-learn: Machine learning in python. J Mach Learn Res 12:2825–2830
Plonka W, Stork C, Šícho M, Kirchmair J (2021) CYPlebrity: machine learning models for the prediction of inhibitors of cytochrome P450 enzymes. Bioorg Med Chem 46:116388
Sun H, Veith H, Xia M, Austin CP, Huang R (2011) Predictive models for cytochrome P450 isozymes based on quantitative high throughput screening data. J Chem Inf Model 51(10):2474–2481
Thölke P, Mantilla-Ramos Y-J, Abdelhedi H, Maschke C, Dehgan A, Harel Y, Kemtur A, Berrada LM, Sahraoui M, Young T et al (2023) Class imbalance should not throw you off balance: choosing the right classifiers and performance metrics for brain decoding with imbalanced data. NeuroImage 277:120253
Xiong Y, Qiao Y, Kihara D, Zhang H-Y, Zhu X, Wei D-Q (2019) Survey of machine learning techniques for prediction of the isoform specificity of cytochrome P450 substrates. Curr Drug Metab 20(3):229–235
Zhao M, Ma J, Li M, Zhang Y, Jiang B, Zhao X, Huai C, Shen L, Zhang N, He L et al (2021) Cytochrome P450 enzymes and drug metabolism in humans. Int J Mol Sci 22(23):12808
Funding
This work was supported in part by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NO. 2020R1A2C2005612) and (NO. 2022R1G1A1004613) and in part by the Korean Big Data Station (K-BDS) with the computing resources, including the technical support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zahid, H., Tayara, H. & Chong, K.T. Harnessing machine learning to predict cytochrome P450 inhibition through molecular properties. Arch Toxicol (2024). https://doi.org/10.1007/s00204-024-03756-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00204-024-03756-9