Abstract
A novel interdomain consortium composed of a methanogenic Archaeon and a sulfate-reducing bacterium was isolated from a microbial biofilm in an oil well in Cahuita National Park, Costa Rica. Both organisms can be grown in pure culture or as stable co-culture. The methanogenic cells were non-motile rods producing CH4 exclusively from H2/CO2. Cells of the sulfate-reducing partner were motile rods forming cell aggregates. They utilized hydrogen, lactate, formate, and pyruvate as electron donors. Electron acceptors were sulfate, thiosulfate, and sulfite. 16S rRNA sequencing revealed 99% gene sequence similarity of strain CaP3V-M-L2AT to Methanobacterium subterraneum and 98.5% of strain CaP3V-S-L1AT to Desulfomicrobium baculatum. Both strains grew from 20 to 42 °C, pH 5.0–7.5, and 0–4% NaCl. Based on our data, type strains CaP3V-M-L2AT (= DSM 113354 T = JCM 39174 T) and CaP3V-S-L1AT (= DSM 113299 T = JCM 39179 T) represent novel species which we name Methanobacterium cahuitense sp. nov. and Desulfomicrobium aggregans sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sulfate-reducing bacteria (SRB) and methanogenic archaea both colonize strictly anoxic biospheres. Herein, the competition for the scarce electron donor H2 is often the driving force for the success of one over the other. While some SRB show a certain resistance toward oxygen (Volbeda et al. 2013) and have an extremely high affinity toward the valuable hydrogen, methanogens lack these characteristics (Kristjansson et al. 1982; Kristjansson and Schönheit 1983; Feldewert et al. 2021; Muyzer and Stams 2008). Therefore, methanogens are often outcompeted in biospheres with limited hydrogen yet sulfate-rich conditions. Interestingly, both, methanogens and sulfate reducers, have been found several times in habitats associated with gas storage facilities or petroleum industry, while sulfate-reducing bacteria are often involved in microbiologically influenced corrosion of the corresponding infrastructure (Volbeda et al. 2013; Mori and Harayama 2011; Molíková et al. 2022; Procópio 2022).
Methane-producing archaea represent a morphologically diverse group within the Euryarchaeota. Most rod-shaped methanogens are assigned to the genera Methanobrevibacter, Methanothermobacter, and Methanobacterium with Methanobacterium as the most diverse group, currently encompassing 24 validly published species. These species were isolated from nearly all over the world, but so far not from Costa Rica. Here, we present the first strain from this area, isolated from a former oil well located very close to the Caribbean Sea.
Most bacterial sulfate reducers cluster within the Deltaproteobacteria. The genera Desulfovibrio and Desulfomicrobium are closely related and were formerly considered as one genus (Rozanova et al. 1988). Herein, Desulfomicrobium species are characterized by the presence of the sulfite reductase desulforubidin and the lack of the enzyme desulfoviridin (Rozanova et al. 1988; Lee et al. 1973). Currently, the genus Desulfomicrobium includes seven validly published species, of which only one is thermophilic.
Our new Desulfomicrobium species (CaP3V-S-L1AT) was isolated from the same biofilm as the Methanobacterium strain (CaP3V-M-L2AT) and both can be grown in pure culture or in a stable co-culture. Therefore, we propose the here described strains CaP3V-M-L2AT and CaP3V-S-L1AT as novel species, Desulfomicrobium aggregans sp. nov. and Methanobacterium cahuitense sp. nov., respectively.
Materials and methods
Sampling and isolation
Both strains were isolated from a natural biofilm, which was extracted from an exploratory oil well in Cahuita National Park, Costa Rica, in September 2016. Sampling technique and location as well as enrichment and cultivation on MS medium were described previously (Dengler et al. 2022).
The initial inoculations were performed using 0.5 mL of environmental sample (containing liquid and natural biofilm particles) in 20 mL medium supplemented with 0.1% acetate (w/v) under H2/CO2 (80:20 v/v, 300 kPa) gas phase. Initial cell growth of methanogenic cells occurred after two weeks of shaking incubation at 37 °C. Here, free methanogenic cells and floating biofilm particles containing both, methanogenic rods and short, non-fluorescent cells, were found. In order to isolate the methanogens, three subsequent dilution series were carried out. All of them failed, and the culture was still contaminated by SRB. To check, whether growth of one of the organisms is dependent on the other, single-cell isolation of both cell types was performed using an optical tweezer for both strains (Huber et al. 1995).
From here on, the medium for the sulfate-reducing strain CaP3V-S-L1AT was changed to MS-Sulf, which equals MS medium but contains an increased sulfate amount of 0.8 g MgSO4 × 7 H2O. For the methanogenic strain, a sulfate-free equivalent was used: SMS medium. Here, all sulfate salts were substituted by equimolar amounts of chloride salts (L−1): 0.45 g NaCl, 5.00 g NaHCO3, 0.083 g MgCl2 × 6 H2O, 0.225 g KH2PO4 × 3 H2O, 0.3 g K2HPO4 × 3 H2O, 0.18 g NH4Cl, 0.06 g CaCl2 × 2 H2O, 0.0016 g NiCl2, 0.0014 g FeCl2, 1 mL 0.1% resazurin solution, 1 mL sulfate-free tenfold trace mineral solution, and 1 mL tenfold vitamin solution (Huber and KO. 2006). Again, both media were supplemented with 0.1% acetate (w/v) and inoculated under H2/CO2 gas atmosphere (80:20 v/v, 300 kPa).
For long-term conservation in our own culture collection, cells were centrifuged under anaerobic conditions (3000 × g, 30 min), re-suspended in their corresponding medium with 5% DMSO. They were then sealed in thin glass capillaries and stored over liquid nitrogen. For short-term storage, logarithmic cell cultures were kept at 4 °C for 2–3 months. Additionally, both strains were deposited within the culture collections of DSMZ and JCM.
For comparative analyses, Methanobacterium subterraneum A8pT (DSM 11,074) and Desulfomicrobium baculatum XT (DSM 4028) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany). Strains were grown in MS-Sulf medium with H2/CO2 gas phase (80:20 v/v, 300 kPa) and 17 mM acetate at their optimal growth temperature.
Phylogenetic analysis
Genomic DNA was isolated using the XS-buffer method (xanthogenate-SDS) (Tillett and Neilan 2000) and 2 mL of exponential cell culture. The 16S rRNA gene was then amplified using the archaeal forward primer 8aF (Eder et al. 1999) and the bacterial forward primer 9bF (Burggraf et al. 1992) together with the universal prokaryotic reverse primer 1512uR (Lane 1991). For the amplification of the mcrA gene, the primer pair MRbac1 (Mori and Harayama 2011) and ME2 (Hales et al. 1996) was used. Then, PCR products were purified using the Wizard® Genomic DNA Purification Kit (Promega GmbH, Walldorf) according to the manufacturer’s instructions. After the clean-up, the PCR product was Sanger-sequenced (LGC Genomics GmbH, Berlin). Gene sequences were surveyed using 4Peaks 1.8 (Griekspoor and Groothuis 2005) and aligned with reference sequences in MEGAX 10.1.8 (Tamura et al. 2013; Kumar et al. 2018) using the ClustalW alignment (Thompson et al. 1994). An approximately maximum-likelihood tree was constructed with FastTree 2 (Price et al. 2010) and visualized using iTol (Letunic and Bork 2007).
The G + C content of the total DNA was defined by genome sequencing. Here, library preparation was carried out compliant with Oxford Nanopore Technologies (ONT, Oxford, the United Kingdom) guidelines for native barcoding of genomic DNA (with EXP-NBD104 and SQK-LSK108). Sequencing was conducted on a MinION MK1C (MinKNOW v.20.10.6). Basecalling and demultiplexing were performed using guppy (fast option, qscore cutoff 7, v. 4.2.3), and reads were assembled with flye (v. 2.8.2) (Kolmogorov et al. 2019). The G + C contents were then determined from the contig sequences in R using the Biostrings package (Pagès et al. 2022).
Morphological and physiological characterization
Gram staining, fluorescence, and phase contrast microscopy were performed as described previously (Dengler et al. 2022). Motility was surveyed for the methanogenic strain CaP3V-M-L2AT at 30–50 °C in two-degree steps under anaerobic conditions using a temperature gradient-forming device (Mora et al. 2014) with a phase contrast microscope (Olympus BX53).
For transmission electron microscopy, exponentially grown cells were chemically fixed with 1% glutaraldehyde (final concentration; v/v) for 10 min at 22 °C and concentrated by centrifugation (4,000 × g, 15 min). 10 µl of cell suspension was placed on copper grids (400-mesh; Plano, Wetzlar, Germany) coated in-house with a 10 nm carbon film, and the samples were subsequently shadowed with Pt/C (15° angle; CFE 50; Cressington). Freeze-etching was performed as described previously (Rachel et al. 2002). Transmission electron micrographs were imaged using a CM12 transmission electron microscope (FEI) operated at 120 keV and fitted with a slow-scan CCD camera (TEM 0124; TVIPS).
For scanning electron microscopy, cells were chemically fixed in cacodylate buffer (50 mM cacodylate, 2 mM MgCl2, and pH 7.0) containing 2.5% (v/v) glutaraldehyde. Then, one drop of the fixed culture was loaded on a microscope glass slide, covered with a large cover slip, and immediately frozen in liquid nitrogen. The coverslip was then broken off and the sample was again treated with glutaraldehyde containing cacodylate buffer. After 15, 30, and 60 min of incubation, the supernatant was removed and replaced with fresh buffer. Then, the samples were contrasted with 1% OsO4 in cacodylate buffer for one hour and dehydrated in a graded series with 10, 20, 40, 60, 80, and 100% acetone for 10 min each. Complete desiccation was achieved by another 20 and 40 min of incubation with 100% acetone. Afterward, critical point drying was executed with liquid carbon dioxide in a critical point dryer (Polaron CAL 9900). Finally, samples were contrasted by sputter-coating with platinum for 40 s (Baltec SCD 050 supercool sputter coater and imaged with a ZEISS Auriga Crossbeam station (ZEISS, Oberkochen, Germany) in the SEM mode via SE-detection at 2 kV acceleration voltage.
Physiological analyses concerning the optimal NaCl concentration, temperature, pH, and substrate specificity were analyzed in triplicates. Sodium chloride was added in concentrations of 0–5% (w/v) and tested in steps of 0.2% between 0 and 1% and in intervals of 0.5% ascending from 1%. In order to determine the substrates used for methanogenesis by strain CaP3V-M-L2AT, the following compounds were tested: acetate (17 mM), formate (22 mM), methanol (31 mM), ethanol (21 mM), 1-propanol (17 mM), 1-butanol (14 mM), 2-propanol (17 mM), 2-butanol (14 mM), methylamine (32 mM), and trimethylamine (17 mM). For the sulfate-reducing strain CaP3V-S-L1AT hydrogen (H2/CO2) lactate (11 mM), formate (22 mM), pyruvate (11 mM), and ethanol (21 mM) were tested as electron donors and sulfate (10 mM), thiosulfate (9 mM), sulfite (1 mM), and elemental sulfur (1% w/v) as electron acceptors. Fermentative growth was tested on fumarate (9 mM), malate (8 mM), lactate (11 mM), pyruvate (11 mM), succinate (9 mM), and propionate (14 mM). Physiological tests were repeated with the addition of 17 mM acetate if growth was not successful after four weeks of incubation to check whether acetate is required for growth. Molarities equal 0.01% (w/v) final concentration in the medium for sulfite and 0.1% (w/v) for all other substrates.
For the determination of optimal growth, cells were counted in triplicates every 24 h for two weeks using a Thoma counting chamber (depth: 0.02 mm). Due to significant biofilm formation, the optimal growth could only be estimated for strain CaP3V-S-L1AT.
In order to examine whether the formation of a stable co-culture is unique to both partners or whether one organism of the community can be substituted by another methanogen or SRB, we performed cross-cultivation experiments with the most closely related species of the novel isolates Methanobacterium subterraneum A8pT and Desulfomicrobium baculatum XT.
Results and discussion
Phylogenetic analysis
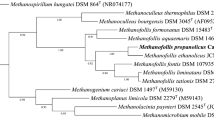
For the methanogenic strain CaP3V-M-L2AT, bidirectional sequencing (LGC Genomics) resulted in a 16S rRNA gene sequence fragment of 1008 b (Fig. 1) and a mcrA gene sequence fragment of 1071 bp (Fig. 2). The phylogenetic analysis revealed that the strain belongs to the genus Methanobacterium. Its closest relative on 16S rRNA gene sequence level was Methanobacterium subterraneum strain A8p with a phylogenetic distance of 0.3%. However, the mcrA gene sequence analysis reveals a very clear position between M. palustre and M. formicicum. The G + C content of the total DNA was 39.3 mol%, which differs significantly from that of M. subterraneum Ap8T (G + C content 54.5 mol%) (Kotelnikova et al. 1998).
The bidirectional sequencing of the sulfate-reducing strain CaP3V-S-L1AT resulted in a 16S rRNA gene sequence fragment of 1415 bp. Here, phylogeny showed that this strain is affiliated with the genus Desulfomicrobium (Fig. 3). The most closely related species were Desulfomicrobium baculatum H.L21 and Desulfomicrobium norvegicum Norway 4 with a phylogenetic distance of 1.48% each. Together with three more species, these closely related species cluster together in the phylogenetic tree. The G + C content of the genomic DNA was 64.5 mol%, which differs significantly from the G + C contents of D. baculatum (56.8 mol%) and D. norvegicum (56.3 mol%) (Rozanova et al. 1988; Sharak Genthner et al. 1997).
Morphological and physiological characterization
Cells of strain CaP3V-M-L2AT showed factor F420 autofluorescence characteristic for methanogens, stained Gram-positive, and were non-motile. In pure culture, rods occurred as single cells or in chains of 2–6 cells with a diameter of 0.2–0.3 μm and a length of 1.4–20 µm. Electron microscopy revealed a thickened cell wall typical for the pseudomurein components in Methanobacterium species and some cells showed cell appendages with a diameter of 5–9 nm (Fig. 4a), reminiscent to fimbriae described for Methanothermobacter thermautotrophicus (Thoma et al. 2008).
a, b Transmission electron micrograph of Pt/C shadowed cells of strain a CaP3V-M-L2AT and b CaP3V-S-L1AT and showing characteristic cell-shapes as well as a fimbriae and b a flagellum. Bars, 1 µm. c, d Scanning electron micrograph of the co-culture showing a densely packed cell aggregate. Bars, 10 µm
Strain CaP3V-M-L2AT used H2/CO2 for methane production, but not acetate, formate, methanol, ethanol, 1-propanol, 1-butanol, 2-propanol, 2-butanol, methylamine, or trimethylamine. Acetate or yeast extract stimulated cell growth on H2/CO2. This stimulating effect of acetate was already described for M. formicicum and M. bryantii (Tab. 1). No effect of added acetate was obtained with the other electron donors. Strain CaP3V-M-L2AT had a doubling time of 6 h under optimal conditions.
Cells of strain CaP3V-S-L1AT were Gram-negative, motile rods. Electron microscopy revealed one polar flagellum with a diameter of 17 nm, typical for the genus Desulfomicrobium (Fig. 4b). In pure culture, the cells rarely occurred as single cells with a diameter of 0.3–0.5 μm and a length of 1–2.5 µm, but mostly formed dense biofilm aggregates with a diameter of up to 1 cm (Fig. 4c, d). This biofilm was formed under all physiological conditions tested so far. Formation of aggregates or biofilm was not observed for the closest relative, Desulfomicrobium baculatum.
Strain CaP3V-S-L1AT used H2, lactate, formate, and pyruvate as electron donors but not ethanol. Electron acceptors were sulfate, thiosulfate, and sulfite but not elemental sulfur. Acetate was required for growth on H2/CO2. Fermentative growth occurred on fumarate and malate but not on pyruvate, lactate, succinate, or propionate. Due to the dense biofilm formation, the doubling time of strain CaP3V-S-L1AT could not be determined reliably.
The growth of both strains CaP3V-M-L2AT and CaP3V-S-L1AT was observed at temperatures ranging from 20 °C to 42 °C. The optimal growth temperature was determined to be 37–40 °C. A pH of 5.0–7.5 supported cell growth and the optimal pH was 5.5–7.5, which is the lowest pH optimum of all Methanobacterium species compared in Table 1. Both strains grew at sodium chloride concentrations from 0 to 4% (w/v) and the optimum was 0–3%.
In co-culture, both strains grew in MS-Sulf medium or MS medium with addition of 10 mM sulfate, 17 mM acetate, and H2/CO2 (80:20) as gas phase. Here, methanogenic rods occurred as planktonic cells or were enclosed in the biofilm of the sulfate reducers (Fig. 4c, d). The SRBs were again densely packed within large cell aggregates. Both, hydrogen sulfide and methane were produced under these conditions. Cross-cultivation experiments with Methanobacterium subterraneum and Desulfomicrobium baculatum indicated that this interdomain consortium is indeed unique. None of our organisms could grow together in co-culture with the corresponding reference strain under the given conditions (i.e., CaP3V-M-L2AT with Desulfomicrobium baculatum and CaP3V-S-L1AT with Methanobacterium subterraneum). D. baculatum outcompeted both methanogens M. subterraneum and the strain CaP3V-M-L2AT, while M. subterraneum outcompeted the novel sulfate-reducing isolate CaP3V-S-L1AT. The co-culture of the novel isolates exists both as an original co-culture received via dilution series and as an artificial co-culture that was later established by newly combining the two isolates.
The former oil well, where both strains were isolated from, displays an open pond with a continuous flow of gas bubbles streaming to the surface. This stream of presumably natural gas might ensure the constant delivery of gaseous nutrients like hydrogen and carbon dioxide. The pond is furthermore heavily influenced by the surrounding rainforest, and large amounts of leaves and organic matter are degraded therein. Photographic material of the pond can be found in the supplements of our previous publication (Dengler et al. 2022). The degradation processes of organic matter in the habitat deliver CO2, acetate, and also further substrates for the fermentative metabolism of the sulfate-reducing isolate. The relatively large sodium chloride range that is tolerated by both organisms could additionally hint on a subsurface connection to the closely located Caribbean Sea (approximate distance: 10–15 m). Such a bridge could also introduce additional sulfates. A drain on the pond enables constant leakage of excess water. The ability to attach to surfaces with cell appendages and to form a biofilm might therefore be extremely useful to stay close to the valuable nutrient influx deriving from the spring.
Taxonomic conclusion
Based on phylogenetics, and morphological and physiological characteristics, the strains CaP3V-M-L2AT and CaP3V-S-L1AT are considered to display novel species within the genera Methanobacterium and Desulfomicrobium, respectively (Table 1, Table 2).
Description of Methanobacterium cahuitense sp. nov.
Methanobacterium cahuitense sp. nov. (ca.hui.ten’se. L. suff. -ense -ensis suffix pertaining to/originating from; N.L. neut. adj. cahuitense originating from Cahuita National Park, Talamanca, Limón, Costa Rica).
Cells are non-motile, rod-shaped, 0.2–0.3 µm in diameter, and 1.4–20 µm in length, occur as single cells or in chains of up two 6 individual cells. Cells stain Gram-positive. Fimbriae are used for adherence. Temperature range for growth is 20–42 °C (optimum, 37–40 °C). Sodium chloride concentration is 0–4% (w/v) (optimum, 0–3%). pH range for growth is 5.0–7.5 (optimum, pH 5.5–7.5). Doubling time is 6 h. H2/CO2 used for methanogenesis. Addition of 0.1% acetate or yeast extract enhances growth. Propanol and butanol inhibit growth. G + C content of DNA is 39.3 mol%.
The type strain is CaP3V-M-L2AT (= DSM 113354 T = JCM 39174 T), isolated from an oil well in the Cahuita National Park, Costa Rica.
Description of Desulfomicrobium aggregans sp. nov.
Desulfomicrobium aggregans sp. nov. (ag’gre.gans. L. part. adj. aggregans adding to, aggregating, forming cell aggregates).
Cells are short rods, 0.3–0.5 µm in diameter, 1–2.5 µm in length, and motile. They occur as single cells or in pairs and always form dense cell aggregates, one polar flagellum, and Gram-negative. Growth temperature is 20–42 °C (optimum, 37–40 °C). Sodium chloride span for growth is 0–40 g/L (w/v) (optimum, 0–30 g/L). pH range is 5.0–7.5 (optimum, pH 5.5–7.5). Electron donors are H2, lactate, formate, and pyruvate, but not ethanol. Electron acceptors are sulfate, thiosulfate, and sulfite, but not elemental sulfur. Acetate is required when H2/CO2 is electron donor. Fermentative growth occurs on fumarate and malate, but not on pyruvate, lactate, succinate, or propionate. G + C content of DNA is 64.5 mol%.
The type strain is CaP3V-S-L1AT (= DSM 113299 T = JCM 39179 T), isolated from an oil well in the Cahuita National Park, Costa Rica.
Data availability
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains CaP3V-M-L2AT and CaP3V-S-L1AT are MW497609 and MW497624 respectively. The corresponding number for the mcrA gene sequence of strain CaP3V-M-L2AT is OP094642.
References
Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43(2):260–296. https://doi.org/10.1016/j.watres.2010.10.010
Burggraf S, Olsen GJ, Stetter KO, Woese CR (1992) A Phylogenetic Analysis of Aquifex pyrophilus. Syst Appl Microbiol 15(3):352–356. https://doi.org/10.1016/S0723-2020(11)80207-9
Cuzin N, Ouattara AS, Labat M, Garcia JL (2001) Methanobacterium congolense sp nov from a methanogenic fermentation of cassava peel. Int J Syst Evol Microbiol 51(2):489–493. https://doi.org/10.1099/00207713-51-2-489
Dengler L, Meier J, Grünberger F, Bellack A, Rachel R, Grohmann D et al (2022) Methanofollis propanolicus sp nov a novel archaeal isolate from a Costa Rican oil well that uses propanol for methane production. Arch Microbiol 204(9):554. https://doi.org/10.1007/s00203-022-03152-w
Dias M, Salvado JC, Monperrus M, Caumette P, Amouroux D, Duran R et al (2008) Characterization of desulfomicrobium salsuginis sp nov and desulfomicrobium aestuarii sp nov two new sulfate-reducing bacteria isolated from the adour estuary (French Atlantic coast) with specific mercury methylation potentials. Syst Appl Microbiol 31(1):30–37. https://doi.org/10.1016/j.syapm.2007.09.002
Eder W, Ludwig W, Huber R (1999) Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep. Red Sea Arch Microbiol 172(4):213–218. https://doi.org/10.1046/j.1529-8817.2000.99079.x
Feldewert C, Lang K, Brune A (2021) The hydrogen threshold of obligately methyl-reducing methanogens. FEMS Microbiol Lett 367(17):1–7. https://doi.org/10.1093/femsle/fnaa137
Genthner BRS, Devereux R (2015) Desulfomicrobium. Bergey’s Man Syst Archaea Bact. https://doi.org/10.1002/9781118960608.gbm01032
Griekspoor A, Groothuis T. 4Peaks, ver. 1.7. Nucleobytes com. 2005;
Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR (1996) Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol 62(2):668–675. https://doi.org/10.12691/jaem-2-4-11
Hippe H, Vainshtein M, Gogotova GI, Stackebrandt E (2003) Reclassification of Desulfobacterium macestii as Desulfomicrobium macestii comb. Nov Int J Syst Evol Microbiol. 53(4):1127–1130. https://doi.org/10.1099/ijs.0.02574-0
Huber R, Burggraf S, Mayer T, Barns SM, Rossnagel P, Stetter KO (1995) Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature 376:57–58. https://doi.org/10.1038/376057a0
Huber, H. Stetter KO. Desulfurococcales. Vol. 3, In The Prokaryotes 3rd eds. New York: Springer; 2006;52–68.
Joulian C, Patel BKC, Ollivier B, Garcia JL, Roger PA (2000) Methanobacterium oryzae sp nov a novel methanogenic rod isolated from a Philippines ricefield. Int J Syst Evol Microbiol 50(2):525–528. https://doi.org/10.1099/00207713-50-2-525
Kolmogorov M, Yuan J, Lin Y, Pevzner PA (2019) Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37(5):540–546. https://doi.org/10.1038/s41587-019-0072-8
König H (1984) Isolation and characterization of Methanobacterium uliginosum sp nov from a marshy soil. Can J Microbiol 30(12):1477–1481. https://doi.org/10.1139/m84-235
Kotelnikova S, Macario AJL, Pedersen K (1998) Methanobacterium subterraneum sp nov a new alkaliphilic, eurythermic and halotolerant methanogen isolated from deep granitic groundwater. Int J Syst Bacteriol 48(2):357–367. https://doi.org/10.1099/00207713-48-2-357
Kristjansson JK, Schönheit P (1983) Why do sulfate-reducing bacteria outcompete methanogenic bacteria for substrates? Oecologia 60(2):264–266. https://doi.org/10.1007/BF00379530
Kristjansson JK, Schönheit P, Thauer RK (1982) Different Ks values for hydrogen of methanogenic bacteria and sulfate reducing bacteria: An explanation for the apparent inhibition of methanogenesis by sulfate. Arch Microbiol 131(3):278–282. https://doi.org/10.1007/BF00405893
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lane DJ. 16S/23S rRNA sequencing. In Nucleic acid techniques in bacterial systematics, eds E. Stackebrandt and M. Goodfellow. Wiley: Chichester
Langendijk PS, Kulik EM, Sandmeier H, Meyer J, van der Hoeven JS (2001) Isolation of desulfomicrobium orale sp nov and desulfovibrio strain NY682, oral sulfate-reducing bacteria involved in human periodontal disease. Int J Syst Evol Microbiol 51(3):1035–1044. https://doi.org/10.1099/00207713-51-3-1035
Lee JP, Yi CS, LeGall J, Peck HD (1973) Isolation of a new pigment, desulforubidin, from Desulfovibrio desulfuricans (Norway strain) and its role in sulfite reduction. J Bacteriol 115(1):453–455. https://doi.org/10.1128/jb.115.1.453-5.1973
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. https://doi.org/10.1093/bioinformatics/btl529
Molíková A, Vítězová M, Vítěz T, Buriánková I, Huber H, Dengler L et al (2022) Underground gas storage as a promising natural methane bioreactor and reservoir. J Energy Storage. 47:138–143
Mora M, Bellack A, Ugele M, Hopf J, Wirth R (2014) The temperature gradient-forming device, an accessory unit for normal light microscopes to study the biology of hyperthermophilic microorganisms. Appl Environ Microbiol 80(15):4764–4770. https://doi.org/10.1128/AEM.00984-14
Mori K, Harayama S (2011) Methanobacterium petrolearium sp nov and Methanobacterium ferruginis sp nov mesophilic methanogens isolated from salty environments. Int J Syst Evol Microbiol 61(1):138–143. https://doi.org/10.1099/ijs.0.022723-0
Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454. https://doi.org/10.1038/nrmicro1892
Pagès H, Aboyoun P, Gentleman R, DebRoy S (2022) Biostrings efficient manipulation of biological strings. R Packag Version. 2640:332
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 approximately maximum-likelihood trees for large alignments. PLoS ONE 5:949
Procópio L (2022) Microbially induced corrosion impacts on the oil industry. Arch Microbiol 204(2):1–6. https://doi.org/10.1007/s00203-022-02755-7
Rachel R, Wyschkony I, Riehl S, Huber H (2002) The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:9–18. https://doi.org/10.1155/2002/307480
Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. Desulfomicrobiaceae. The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. 2014;9783642390:1–413. Doi: https://doi.org/10.1007/978-3-642-39044-9
Rozanova E, Nazina T, Galushko A (1988) Isolation of a new genus of sulfate-reducing bacteria and description of a new species of this genus Desulfomicrobium apsheronum gen nov sp nov. Mikrobiologiya 57:634–641
Sharak Genthner BR, Friedman SD, Devereux R (1997) Reclassification of Desulfovibrio desulfuricans Norway 4 as Desulfomicrobium norvegicum comb nov and confirmation of Desulfomicrobium escambiense as a new species in the genus Desulfomicrobium. Int J Syst Bacteriol 47(3):889–892. https://doi.org/10.1099/00207713-47-3-889
Shlimon AG, Friedrich MW, Niemann H, Ramsing NB, Finster K (2004) Methanobacterium aarhusense sp nov a novel methanogen isolated from a marine sediment (Aarhus Bay, Denmark). Int J Syst Evol Microbiol 54(3):759–763. https://doi.org/10.1099/ijs.0.02994-0
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Thoma C, Frank M, Rachel R, Schmid S, Näther D, Wanner G, Wirth R (2008) The Mth60 fimbriae of Methanothermobacter thermoautotrophicus are functional adhesins. Env Microbiol 10(10):2785–2795. https://doi.org/10.1111/j.1462-2920.2008.01698.x
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
Tillett D, Neilan BA (2000) Xanthogenate nucleic acid isolation from cultured and environmental Cyanobacteria. J Phycol 36(1):251–258. https://doi.org/10.1046/j.1529-8817.2000.99079.x
Volbeda A, Amara P, Iannello M, De Lacey AL, Cavazza C, Fontecilla-Camps JC (2013) Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase. Chem Commun 49:7061–7063. https://doi.org/10.1039/c3cc43619e
Worakit S, Boone DR, Mah RA, Abdel-Samie M-E, El-Halwagi MM (1986) Methanobacterium alcaliphilum sp nov an H2-utilizing methanogen. Int J Syst Bacteriol 36(3):380–382. https://doi.org/10.1099/00207713-36-3-380
Zellner G, Bleicher K, Braun E, Kneifel H, Tindall BJ, de Macario EC et al (1988) Characterization of a new mesophilic, secondary alcohol-utilizing methanogen, Methanobacterium palustre spec nov from a peat bog. Arch Microbiol 151(1):1–9. https://doi.org/10.1111/j.1574-6968.1987.tb02309.x
Acknowledgements
We thank Felix Grünberger for the determination of G + C content. Furthermore, we thank all team members within the Costa Rican offices at CONAGEBIO and SINAC-ACLAC-MINAE for their dedicated support and cooperation, especially Melania Muñoz García, Angela González Grau, and Jorge Arturo Gonzáles Villalobos. Permit no. SINAC-ACLAC-PIME-R-030-2016 and R-044-2016-OT-CONAGEBIO.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by funds provided by the University of Regensburg to D.G.
Author information
Authors and Affiliations
Contributions
LD and HH conceived the study. LD and JM wrote the manuscript and performed the morphological, physiological and 16S rRNA analyses. AK performed SEM. LD, RR, LN performed TEM. DG designed the G + C analysis. LD, HH, JM, AB, AK and RR prepared figures. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dengler, L., Meier, J., Klingl, A. et al. A novel interdomain consortium from a Costa Rican oil well composed of Methanobacterium cahuitense sp. nov. and Desulfomicrobium aggregans sp. nov.. Arch Microbiol 205, 189 (2023). https://doi.org/10.1007/s00203-023-03533-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03533-9