Abstract
A novel methanogenic strain, CaP3V-MF-L2AT, was isolated from an exploratory oil well from Cahuita National Park, Costa Rica. The cells were irregular cocci, 0.8–1.8 μm in diameter, stained Gram-negative and were motile. The strain utilized H2/CO2, formate and the primary and secondary alcohols 1-propanol and 2-propanol for methanogenesis, but not acetate, methanol, ethanol, 1-butanol or 2-butanol. Acetate was required as carbon source. The novel isolate grew at 25–40 °C, pH 6.0–7.5 and 0–2.5% (w/v) NaCl. 16S rRNA gene sequence analysis revealed that the strain is affiliated to the genus Methanofollis. It shows 98.8% sequence similarity to its closest relative Methanofollis ethanolicus. The G + C content is 60.1 mol%. Based on the data presented here type strain CaP3V-MF-L2AT (= DSM 113321T = JCM 39176T) represents a novel species, Methanofollis propanolicus sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Methanofollis (Zellner et al. 1999) represents one out of six genera within the family Methanomicrobiaceae (Balch et al. 1979). With the publication of this genus, the former Methanogenium species M. tationis and M. liminatans were transferred to the novel genus Methanofollis due to distinct patterns of glycolipids, phosphoglycolipids and amino-phosphoglycolipids among others. Furthermore, Methanofollis tationis possesses a pterin that is different from the known methanopterin or sarcinapterin–tatiopterin has not been found in any other methanogen since then (Raemakers-Franken et al. 1991).
Today, six validly described species are assigned to the genus Methanofollis. These species were isolated from various aquatic environments with different salinities. Methanofollis liminatans was derived from a wastewater reactor in Germany and Methanofollis tationis from a solfataric field in Chile. Methanofollis formosanus (Wu et al. 2005) and Methanofollis aquaemaris (Lai and Chen 2001) were both isolated from a fish pond in Taiwan. Methanofollis ethanolicus (Imachi et al. 2009) was discovered in a lotus field, while the recently described Methanofollis fontis was isolated from a marine sediment near a cold seep (Chen et al. 2020). With the increasing number of described species, it became clear that the genus’ capability to use primary and secondary alcohols with two or more C-atoms for methane production is a characteristic feature.
CaP3V-MF-L2AT shows a similarity of 98.8% in 16S rRNA gene sequence towards Methanofollis ethanolicus; however, their physiological characteristics differ significantly. Strain CaP3V-MF-L2AT cannot grow on ethanol or 1-butanol but utilizes 1-propanol and 2-propanol for methanogenesis. Furthermore, strain CaP3V-MF-L2AT is characterized by a smaller cell size, its motility and a significantly shorter generation time. Therefore, we propose the here described strain CaP3V-MF-L2AT as a novel species, Methanofollis propanolicus sp. nov.
Materials and methods
Sampling and isolation
Strain CaP3V-MF-L2AT was isolated from an exploratory oil well in Cahuita National Park, located at the southwestern Atlantic coast of Costa Rica (SI Fig. 1). Water samples were taken under sterile and anaerobic conditions in a depth of 20–30 cm sub-watersurface using 100 ml glass bottles.
MS medium, modified from Balch’s medium I (Balch et al. 1979) was used for enrichment and cultivation of strain CaP3V-MF-L2AT. This medium contained the following components (l−1): 0.45 g NaCl, 5.00 g NaHCO3, 0.1 g MgSO4 × 7 H2O, 0.225 g KH2PO4 × 3 H2O, 0.3 g K2HPO4 × 3 H2O, 0.025 g (NH4)2SO4, 0.06 g CaCl2 × 2 H2O, 0.002 g (NH4)2Ni(SO4)2, 0.002 g FeSO4 × 7 H2O, 1 ml 0.1% resazurin solution, 1 ml tenfold trace mineral solution and 1 ml tenfold vitamin solution (Huber and Stetter 2006). The medium was prepared according to the standard techniques for anaerobic cultivation (Balch and Wolfe 1976). It was reduced with 0.5 g Na2S × 2–3 H2O and the pH was adjusted to 6.5 with 1 M HCl. 20 ml medium were aliquoted into 120 ml serum bottles and pressurized with H2/CO2 (80:20, v/v, 300 kPa) or N2/CO2 (80:20, v/v, 200 kPa), respectively.
Inoculation was performed with 0.5 ml of original samples in 20 ml medium supplemented with 0.5% acetate (w/v) and H2/CO2 as gas phase. Initial cell growth with irregular cocci occurred after 1 week of incubation at 37 °C with shaking. After two transfers, single cell isolation was performed using an optical tweezer setup (Huber et al. 1995). For further studies, acetate supplement was reduced from 0.5 to 0.1%. For long-term conservation, cells were anaerobically centrifuged (3000× g, 30 min), resuspended in medium containing 5% DMSO, sealed in glass capillaries and stored over liquid nitrogen in our in-house culture collection. Logarithmic cell cultures were stored at 4 °C for 2–3 months for short-term storage.
Microscopic techniques
Light microscopy was performed using a phase-contrast and fluorescence microscope (Olympus BX60; Bright Line HC 434/17 excitation filter with a beam splitter 452 nm and long pass filter U-E455). Motility was surveyed at 37 °C under anaerobic conditions using a temperature gradient-forming device (Mora et al. 2014) connected to a phase-contrast microscope (Olympus BX53). Gram-stain was carried out as described previously (Boone and Whitman 1988) using Escherichia coli K12 (ATCC® 25922™) and Bacillus atrophaeus (ATCC® 9372™) as reference strains.
For electron microscopy, cells in late exponential growth phase were fixed with 1% glutardialdehyde (final concentration; v/v) for 10 min at room temperature and concentrated by centrifugation (4000× g, 15 min). 10 µl of cell concentrate were placed on hydrophilized 400-mesh carbon-coated copper grids (Plano). Samples on grids were then either negatively stained for 1 min with 2% uranyl acetate (w/v) or shadowed with Pt/C (15° angle; CFE 50; Cressington). Freeze etching was carried out as described previously (Rachel et al. 2002).
Transmission electron micrographs were imaged using a CM12 transmission electron microscope (FEI) operated at 120 keV and equipped with a slow-scan charge-coupled device camera (TEM 0124; TVIPS).
Phylogenetic analysis
DNA isolation was carried out using the XS-buffer method (xanthogenate-SDS) (Tillett and Neilan 2000) using 2 ml of late exponential cell culture as starting material. The 16S rRNA gene was PCR-amplified using the universal archaeal forward primer 8aF (Eder et al. 1999) and the universal microbial reverse primer 1512uR (Lane 1991). Subsequently, DNA was purified using Wizard® Genomic DNA Purification Kit (Promega) according to the manufacturers’ instructions. Bidirectinoal Sanger sequencing was carried out by LGC Genomics GmbH, Berlin. Sequences were manually revised using 4Peaks 1.8 (Griekspoor and Groothuis 2005) and aligned with 16S rRNA gene sequences of type strains within the family Methanomicrobiaceae in MEGAX 10.1.8 (Tamura et al. 2013; Kumar et al. 2018) using the ClustalW alignment (Thompson et al. 1994). An approximately maximum-likelihood tree was constructed applying FastTree 2 (Price et al. 2010) and visualized using iTol (Letunic and Bork 2007).
The G + C content of total DNA was determined by genome sequencing. Therefore, library preparation was performed according to Oxford Nanopore Technologies (ONT, Oxford, United Kingdom) protocol for native barcoding of genomic DNA (with EXP-NBD104 and SQK-LSK109). Sequencing was performed on ONT’s MinION MK1C device (MinKNOW v.20.10.6). After basecalling and demultiplexing using guppy (fast option, qscore cutoff 7, v. 4.2.3), reads were assembled using flye (v. 2.8.2) (Kolmogorov et al. 2019). Finally, the G + C content was calculated from the contig sequences using the Biostrings package in R (Pagès et al. 2022).
Morphological and physiological characterization
The analysis of physiological parameters such as optimal temperature, pH, NaCl, and substrate specificity were carried out in triplicates. Experiments on the pH spectrum were conducted by adjusting the values at room temperature using anaerobic HCl or NaOH, respectively. To assure constant pH values, pH was controlled over the time of incubation and readjusted if necessary. To study sodium chloride requirements, the salt was added in concentrations of 0–4.0% (w/v) to NaCl-free MS medium. Sodium chloride was tested in intervals of 0.2% between 0 and 1% and in steps of 0.5% ascending from 1%. To determine substrates used for methanogenesis, the following compounds were added to the medium: acetate (17 mM), formate (22 mM), methanol (31 mM), ethanol (22 mM), 1-propanol (17 mM), 1-butanol (14 mM), 2-propanol (17 mM) and 2-butanol (14 mM). Molarities equal 0.1% (v/v) final concentration in the culture medium. In addition, 5 mM ethanol and 0.01% yeast extract were supplemented as described for Methanofollis ethanolicus (Imachi et al. 2009). For the calculation of optimal growth, cells were counted every 24 h for 1 week using a Thoma counting chamber (0.02 mm depth).
Results and discussion
Phylogenetic analysis
Bidirectional sequencing (LGC Genomics) resulted in a 16S rRNA gene sequence fragment of 1364 bp. Phylogenetic analysis showed that strain CaP3V-MF-L2AT is affiliated to the genus Methanofollis (Fig. 1). Its closest relative is Methanofollis ethanolicus HASUT with a phylogenetic distance of 1.2%. The G + C content was 60.1 mol%.
Morphological and physiological characterization
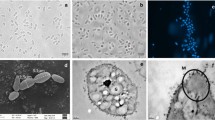
Cells of strain CaP3V-MF-L2AT showed a greenish autofluorescence of factor F420 typically found in methanogens, stained Gram-negative and were motile. In continuous culture, irregularly shaped cocci with a diameter of 0.8–1.8 μm occurred singly or in pairs. Electron microscopy not only revealed an uneven cell surface but also highly variable shape ranging from conical to circularly dent cells (Fig. 2). Cells showed two different types of cell appendages: archaella with a diameter of 12 nm and pili with a diameter of 8 nm. Cells exhibit the typical S-Layer structure of Methanofollis species (Fig. 3).
Growth of strain CaP3V-MF-L2AT was detected from 20 to 40 °C, with an optimal growth temperature of 37 °C. A pH of 6.0–7.5 supported cell growth, while levels below or above did not. The optimal pH was determined at pH 6.5–7.0. Strain CaP3V-MF-L2AT tolerated sodium chloride concentrations from 0 to 2.5% (w/v), while the optimal range was 0.2–1.5%.
Strain CaP3V-MF-L2AT used H2/CO2, formate, 1-propanol, and 2-propanol as energy source, while acetate was required as a carbon source. This substrate spectrum differed from all other described species of the genus Methanofollis (Table 1). Yeast extract had a stimulating effect on cell growth as described for M. ethanolicus and M. liminatans. Acetate as well as the primary and secondary alcohols methanol, ethanol, 1-butanol, 2-butanol, and cyclopentanol did not support cell growth when used alone or in combination with acetate or yeast extract. This was even true for the substrate combination of 5 mM ethanol and 0.01% yeast extract with an incubation time of up to 3 months (which works for Methanofollis ethanolicus). The doubling time of strain CaP3V-MF-L2AT under optimal physiological conditions was 16 h.
The ancient oil well, where strain CaP3V-MF-L2AT was isolated from, represents an open pond with plenty of organic import (e.g., leaves from surrounding fauna). This explains the availability of substrates, such as acetate or propanol. In the natural environment, propanol derives from anaerobic microbial degradation processes. 2-propanol is known to be produced by some saccharolytic Clostridia by the reduction of acetone (Langlykke et al. 1937; Kutzenok and Aschner 1952; George et al. 1983).
Taxonomic conclusion
Based on phylogenetic, morphological, and physiological characteristics, strain CaP3V-MF-L2AT is considered to display a novel species within the genus Methanofollis (Table 1).
Description of Methanofollis propanolicus sp. nov.
Methanofollis propanolicus sp. nov. (pro.pa.no´li.cus. N.L. n. propanol; L. suf. -icus -a -um suffix used with various meanings; N.L. masc. adj. propanolicus) regarding to propanol, based on the substrate propanol, which can be metabolized by this species.
Cells are irregular cocci, motile, 0.8–1.8 μm in diameter, and occur as single cells or in pairs. Occurrence of at least two types of cell appendages. Strictly anaerobic. Temperature range for growth 25 °C–40 °C (optimum, 37 °C). Sodium chloride range for growth 0–2.5% (w/v) (optimum, 0.2–1.5%). pH range for growth 6.0–7.7 (optimum, pH 6.5–7.0). Doubling time is 16 h. H2/CO2, formate, 1-propanol and 2-propanol used for methanogenesis, addition of 0.1% acetate is crucial for growth on substrates different from 1-propanol or 2-propanol. G + C content of DNA is 60.1 mol%. Closely related to Methanofollis ethanolicus JCM15103T (98.8% 16S rRNA gene sequence similarity).
The type strain is CaP3V-MF-L2AT (= DSM 113321 T = JCM 39176 T), isolated from an oil well in the Cahuita National Park, Costa Rica.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain CaP3V-MF-L2AT is MW490723.
References
Balch WE, Wolfe RS (1976) New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol 32(6):781–791
Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS (1979) Methanogens: reevaluation of a unique biological group. Microbiol Rev 43(2):260–296. https://doi.org/10.1016/j.watres.2010.10.010
Boone DR, Whitman WB (1988) Proposal of minimal standards for describing new taxa of methanogenic bacteria. Int J Syst Bacteriol 38(2):212–219
Chen S-C, Teng N-H, Lin Y-S, Lai M-C, Chen H-H, Wang C-C (2020) Methanofollis fontis sp. nov., a methanogen isolated from marine sediment near a cold seep at four-way closure ridge offshore southwestern Taiwan. Int J Syst Evol Microbiol 70(10):5497–5502. https://doi.org/10.1099/ijsem.0.004440
Eder W, Ludwig W, Huber R (1999) Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep. Red Sea Arch Microbiol 172(4):213–218
George HA, Johnson JL, Moore WEC, Holdeman LV, Chen JS (1983) Acetone, isopropanol, and butanol production by Clostridium beijerinckii (syn. Clostridium butylicum) and Clostridium aurantibutyricum. Appl Environ Microbiol 45(3):1160LP-1163LP
Griekspoor A, Groothuis T (2005) 4Peaks, ver. 1.7. Nucleobytes com.
Huber H, Stetter KO (2006) Desulfurococcales, 3rd edition. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes, 3rd edn. Springer, New York, pp 52–68
Huber R, Burggraf S, Mayer T, Barns SM, Rossnagel P, Stetter KO (1995) Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature 376:57–58
Imachi H, Sakai S, Nagai H, Yamaguchi T, Takai K (2009) Methanofollis ethanolicus sp nov, an ethanol-utilizing methanogen isolated from a lotus field. Int J Syst Evol Microbiol. 59(4):800–805. https://doi.org/10.1099/ijs.0.003731-0
Kolmogorov M, Yuan J, Lin Y, Pevzner PA (2019) Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37(5):540–546. https://doi.org/10.1038/s41587-019-0072-8
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Kutzenok A, Aschner M (1952) Degenerative processes in a strain of Clostridium butylicum. J Bacteriol 64(6):829–836. https://doi.org/10.1128/JB.64.6.829-836.1952
Lai MC, Chen SC (2001) Methanofollis aquaemaris sp. nov., a methanogen isolated from an aquaculture fish pond. Int J Syst Evol Microbiol 51(5):1873–1880
Lane DJ (1991) 16S23S rRNA sequencing. Nucleic acid Tech Bact Syst. 15–175.
Langlykke AF, Peterson WH, Fred EB (1937) Reductive processes of Clostridium butylicum and the mechanism of formation of isopropyl alcohol. J Bacteriol 34(4):443–453. https://doi.org/10.1128/JB.34.4.443-453.1937
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. https://doi.org/10.1093/bioinformatics/btl529
Mora M, Bellack A, Ugele M, Hopf J, Wirth R (2014) The temperature gradient-forming device, an accessory unit for normal light microscopes to study the biology of hyperthermophilic microorganisms. Appl Environ Microbiol 80(15):4764–4770
Pagès H, Aboyoun P, Gentleman R, DebRoy S (2022) Biostrings: efficient manipulation of biological strings. R Packag Version 2640. https://doi.org/10.18129/B9.bioc.Biostrings
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):e9490
Rachel R, Wyschkony I, Riehl S, Huber H (2002) The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:307480. https://doi.org/10.1155/2002/307480
Raemakers-Franken PC, van Elderen CH, Van der Drift C, Vogels GD (1991) Identification of a novel tatiopterin derivative in Methanogenium tationis. BioFactors 3(2):127–130
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tillett D, Neilan BA (2000) Xanthogenate nucleic acid isolation from cultured and environmental Cyanobacteria. J Phycol 36(1):251–258. https://doi.org/10.1046/j.1529-8817.2000.99079.x
Wu SY, Chen SC, Lai MC (2005) Methanofollis formosanus sp. nov., isolated from a fish pond. Int J Syst Evol Microbiol 55(2):837–842
Zabel HP, König H, Winter J (1984) Isolation and characterization of a new coccoid methanogen, Methanogenium tatii, new species from a solfataric field on Mount Tatio (Chile). Arch Microbiol 137:308–315
Zellner G, Sleytr UB, Messner P, Kneifel H, Winter J (1990) Methanogenium liminatans, spec. nov., a new coccoid,mesophilic methanogen able to oxidize secondary alcohols. Arch Microbiol 153:287–293
Zellner G, Boone DR (2001) Genus III. Methanofollis Zellner, Boone, Keswani, Whitman, Woese, Hagelstein, Tindall and Stackebrandt 1999, 253VP. In: Boone DR, Castenholz RW, Garrity GM (eds) Bergey’s manual of systematic bacteriology, the Archaea and the deeply branching and phototrophic Bacteria, vol 1, 2nd edn. Springer, New York, pp 253–255
Zellner G, Boone DR, Keswani J, Whitman WB, Woese CR, Hagelstein A et al (1999) Reclassification of Methanogenium tationis and Methanogenium liminatans as Methanofollis tationis gen. nov., comb. nov. and Methanofollis liminatans comb. nov. and description of a new strain of Methanofollis liminatans. Int J Syst Bacteriol 49(1):247–255. https://doi.org/10.1099/00207713-49-1-247
Acknowledgements
We thank Daniel Eckl for providing bacterial reference strains and the Costa Rican authorities CONAGEBIO and SINAC-ACLAC–MINAE for long-term support and fruitful collaboration.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work received no specific grant from any funding agency, but was supported by funds provided by the University of Regensburg to DG.
Author information
Authors and Affiliations
Contributions
LD and HH conceived the study. LD and JM wrote the manuscript and performed the morphological, physiological and 16S rRNA analyses. DG designed and FG performed the G + C analysis. LD, HH, JM, AB and RR prepared figures. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

203_2022_3152_MOESM1_ESM.jpg
SI Fig 1: Ancient oil well in the Cahuita National Park, Costa Rica, original sampling site of strain CaP3V-MF-L2AT Supplementary file1 (JPG 68748 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dengler, L., Meier, J., Grünberger, F. et al. Methanofollis propanolicus sp. nov., a novel archaeal isolate from a Costa Rican oil well that uses propanol for methane production. Arch Microbiol 204, 554 (2022). https://doi.org/10.1007/s00203-022-03152-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03152-w