Abstract
This study was designed to evaluate the antimicrobial, antioxidant, and cytotoxic potentials of the marine actinomycetes spp. isolated from the Red Sea water, Hurghada, Egypt. Out of 80 actinomycetes isolates, one isolate AW6 was selected based on its antioxidant activity (IC50 about 5.24 µg/mL which scavenged 91% of formed DPPH free radicals) and antimicrobial potential against E. coli, S. aureus, B. subtilis, and P. aeruginosa, A. niger, and C. albicans. The strain was identified based on phenotypic and genotypic analysis, and deposited in the GenBank with accession number OK090864.1. Cultivation of the selected strain on rice, chromatographic purification, and structural elucidation led to the isolation of two compounds C1: umbelliferone, and C2: 1-methoxy-3-methyl-8-hydroxy-anthraquinone. The antimicrobial activity of the obtained compounds showed that C1 and C2 have low antibacterial activity toward S. aureus and E. coli with no pronounced activity toward P. aeruginosa, C. albicans, and A. niger. Additionally, the antioxidant activity of C1 and C2 revealed that C2 has a good antioxidant activity, with DPPH scavenging activity reaching (55.25%), followed by C1 (30.20%). Moreover, both compounds displayed anti-Gyr-B enzyme activity with IC50 value of (3.79 ± 0.21 µM) for C1, and (IC50 = 13 ± 0.71 µM) for C2. The ADME-related physicochemical properties of the obtained compound were predicted using SwissADME web tools and the ProToxii webserver was used to estimate in silico toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Actinomycetes are the most abundant category of microorganisms in nature, primarily found in soil and marine water (Abdel-Aziz et al. 2018). They have produced a large number of essential bioactive chemicals with great economic value and are still constantly tested for the production of new bioactive molecules. Actinomycetes contain around two-thirds of naturally occurring antibiotics, including several medically significant compounds (Palazzotto et al. 2019). Almost 80% of the world’s antibiotics are known to be derived from Actinomycetes, primarily the species Streptomyces and Micromonospora (van der Meij et al. 2017). Most antibiotics are no longer effective against the majority of bacteria-caused infections (Ghareeb et al. 2015; Elkhouly et al. 2021a). Staphylococcus aureus is a well-known organism that causes illnesses such as pneumonia and recently has gained resistance to the majority of antibiotic groups (Tong et al. 2015; Hamed et al. 2020a, b; El-Shazly et al. 2021). During this time, vancomycin has been the therapeutic antibiotic against Methicillin-resistant Staphylococcus aureus (MRSA), but unfortunately, Vancomycin-resistant strains have emerged clinically (Gardete and Tomasz 2014). Vancomycin-resistant S. aureus (VRSA) is considered a critical problem to doctors not only because of its vancomycin and methicillin resistance but also due to resistance to a variety of other antibiotics, including aminoglycosides, fluoroquinolones, and macrolides (Kest and Kaushik 2019). Certain unfavorable side effects and the spread of diseases associated with this novel antimicrobial medication resistance highlight the necessity for the development of alternative newer antimicrobial medicines with effectiveness against Gram-positive bacteria (Mohammed et al. 2019; Elkhouly et al. 2021b). Furthermore, Gram-negative antibiotic-resistant opportunistic bacteria with a multi-drug resistance threaten patients in hospitals and communities as they possess a widespread resistance to the first, second, and third generations of penicillin and cephalosporin (Mohammed et al. 2019). These bacteria, such as Pseudomonas aeruginosa, are widespread organisms found in the environment that operate as opportunistic pathogens in clinical circumstances where the patient’s defense system is impaired (Moradali et al. 2017). As a result of this phenomenon, many strains of bacteria have developed antibiotic resistance, and in many cases, multi-resistant, to these known therapeutic medicines, and to address this issue, a new antibiotic with a novel mechanism of action is required (Ventola 2015).
The present study is carried out to explore the role of Actinomycetes compounds as antioxidant and antimicrobial agents against different types of pathogens including P. aeruginosa, S. aureus, E. coli, A. niger, and C. albicans. The anti-Gyr activity of the isolated compounds was also investigated. Furthermore, the ADME-related physicochemical properties of the obtained compound were predicted using SwissADME web tools and the ProToxii webserver was used to estimate in silico toxicity.
Materials and methods
Collection of sediment samples
In June 2018, eight marine samples were obtained from the Red Sea at depths 10, 15. 32 and 40 m using a sterile core sampler. The sediment samples were collected and serially-numbered before being transported to the Microbiology laboratory and kept at 4 °C at the National Research Centre.
Isolation of actinomycetes from sediment samples
The isolation of marine actinomycetes was performed using a starch nitrate agar medium containing 50% salt water and the following ingredients: (g/L): 20 starch; 0.5 K2HPO4; 1 KNO3; 0.5 MgSO4 7H2O; 0.01 FeSO4; and 15 agar. Before sterilization, the pH of the medium was adjusted to 7, and Mycostatin (500 L/L) was added afterward. 1 g of sediment was taken from each sample and mixed separately with 9 mL of sterile saltwater. In 100 mL of sterile starch nitrate agar medium, five milliliters of the suspension were added. Each sterilized plate received approximately 20 mL of medium with no air bubbles. The inoculated plates were incubated for 7 days at 30 °C. After incubation, the actinomycete colonies were picked and purified using the streaking method. The purified strains were preserved on starch nitrate slants and 25% glycerol stocks at − 20 °C (Burhamzah et al. 2016).
Small-scale fermentation and screening of antimicrobial activity
Each isolate was grown on rice substrate, extracted with ethyl acetate, and evaporated to test its antimicrobial activity. The agar well diffusion method was used to test the antimicrobial activity of crude actinomycetes extracts. Sterile Mueller Hinton agar plates were swabbed separately with the pathogens as test microbes including Gram-positive bacteria; Bacillus subtilis (ATCC66), Staphylococcus aureus (ATCC6538-P), and Methicillin-resistant Staphylococcus aureus (MRSA) (ATCC25923); Gram-negative bacteria Escherichia coli (ATCC14169), Pseudomonas aeruginosa (ATCC 27853), Klebsiella pneumoniae, Salmonella typhi, and Salmonella enterica; yeasts Candida albicans (ATCC10231), and (ATCC9080) and fungus Aspergillus niger (NRRL A-326). In each plate, 6 mm diameter wells were drilled with a sterile cork borer and approximately 20 µL of each extract was put into each well against each of the test organisms. After that, plates were incubated for 24 h at 37 °C. The diameter of the inhibition zones was used to determine antimicrobial activity. Only isolates with broad-spectrum activity were chosen for further study (Waithaka et al. 2017). Antimicrobial activity was observed after 24 h of incubation at 37 °C for bacteria and 48 h of incubation at 25 °C for fungus, and inhibition zones were reported as diameters (mm).
Identification of the most potent isolate
Phenotypic identification
Morphological methods, including macroscopic and microscopic ones, were used to characterize the potent isolate up to the genus level. Through oil immersion (100X), the mycelium structure, color, and arrangement of conidiophores were observed. The studying of spore-bearing hyphae, spore chain structure, spore color, aerial mass color, and substrate mycelia color were studied according to Bergey (1989) and the International Streptomyces Project (ISP). Additionally, morphological and cultural characteristics of the potent isolate AW6 were studied in different media following the instructions given by the International Streptomyces Project (ISP) (Singh et al. 2012). The biochemical tests were performed by assessing the oxidase enzyme ability, indole test, methyl red test, citrate utilization ability, carbohydrate fermentation, and hydrogen sulfide production nitrate reduction (Pradhan and Tamang 2019).
Genotypic identification
Based on the antimicrobial activity of the crude extracts, one isolate coded AW6 was genetically identified by sequencing the isolate’s 16S rRNA gene. The Qiagen DNeasy Blood & Tissue Kit (Thermo Fisher Scientific, Waltham, USA) was used to extract the DNA, which was done according to the manufacturer’s recommendations. Two universal primers were used in the PCR amplification (27F5′-AGAGTTTGATCCTGGCTCAG-3′; 1492R 5′-GGTTACCTTGTTACGACTT-3′). The final volume of the PCR amplification reaction was as follows: 50 μL (5 μL of 10 × Dream Taq Green PCR buffer, 2 μL of each 10 μmol dm−3 primers, 5 μL of 2 mmol dm−3 dNTP, 0.3 μL Taq DNA polymerase, and 0.5 μL of template DNA). The PCR reaction ran under the following conditions: 94 °C for 45 s, 55 °C for 60 s, and 72 °C for 60 s. The purified product was sequenced at Macrogen Company, South Korea. The similarity and homology of the 16S rDNA sequences were investigated by comparing the obtained sequences with similar known sequences in the NCBI database using online BLAST alignment search tools (http://www.ncbi.nlm.nih.gov/BLAST). MEGA-X software was used to create the phylogenetic tree (Kumar et al. 2016).
Fermentation and production of bioactive compounds
Rice in the solid state was used as a fermentation medium; the selected isolate was grown into a 250 mL Erlenmeyer flask containing 50 mL starch nitrate broth (pH 7) at 30 °C for 7 days under shaking conditions. After incubation, the starch broth culture medium was used as a seed culture to inoculate the rice media for large-scale fermentation (Hamed et al. 2022).
Extraction of bioactive compounds
Extraction of the bioactive metabolites from the fermented rice medium inoculated with Actinomyces sp. AW6 was carried out using an equal volume of ethyl acetate after shaking for one hour for complete extraction (Hamed et al. 2022). The ethyl acetate was separated and evaporated at 40 °C to obtain 13 g of the dry extract.
Purification of bioactive secondary metabolites
Purification and structure elucidation were carried out initially using flash column chromatography. 10 g from the obtained crude extract was applied on a 7 cm diameter column filled with normal phase silica. The ratio of 20:1 adsorbent (silica gel) to solute (crude extract). A total of 100 fractions of 10 mL each were collected and analyzed using thin-layer chromatography (TLC) to identify the fractions with compounds of interest. The most potent fraction was further purified using the size-exclusion chromatography technique and performed using Sephadex LH-20 sub-column. The separation was based on molecular weight. The purified compounds were subjected to different spectroscopic analyses such as NMR and ESI for structure elucidation.
Biological activity evaluation
Antimicrobial activity
The antimicrobial activity measurement of Actinomyces sp. AW6 compounds was carried out using an antimicrobial assay and MIC as described by Hamed et al. (2020a, b), Alhadrami et al. (2021), and Qader et al. (2021). All test pathogens were obtained from the Culture Collection Center (Microbial Chemistry Department and National Research Centre, NRC), Egypt.
Determination of free radical scavenging activity (RSA)
The free radical scavenging activity (RSA) was measured by the decoloration of an ethanolic solution of DPPH radical spectrophotometrically at 517 nm following Brand-Williams et al (1995) technique. The scavenging activity was calculated as follows:
In vitro DNA gyrase-B inhibition
The DNA gyrase-B and ParE inhibitory activities were determined using the Inspiralis assay kit (Inspiralis®, London, UK) on streptavidin-coated 96-well microtiter plates (Thermo Scientific, Hamburg, Germany), according to the manufacturer’s protocol (Durcik et al. 2018). The experiment measures the capacity of the separated drugs to inhibit the ATPase activity of both the gyrase-B and ParE subunits.
In silico predictions ADME-related physicochemical properties
The ADME-related physicochemical properties of the obtained compound were predicted using SwissADME web tools (Daina et al. 2017).
In silico toxicity prediction
The in silico prediction of toxicity for compounds was performed via ProTox ii web server as previously reported (Banerjee et al. 2018).
Results and discussion
Isolation of endophytic actinobacteria
Using serial dilution technique, several actinobacterial strains were isolated from collected marine samples, i.e., algal homogenate, marine sediment, or marine water. Several studies have isolated many actinomycetes isolates from marine samples, and the isolation was depended on visualization of actinomycetes colonies on specific media for actinomycetes (Hamed et al. 2022), Table 1 summarizes the number of isolates and their sources.
Antimicrobial screening for isolated endophytic actinobacteria
Eighty isolates were selected based on morphological examination as actinomycetes and screened biologically for their antimicrobial activity. Based on the antimicrobial activity of the tested strains, the isolate AW6 has been selected for further investigation Table 2 shows the antimicrobial activity of some selected actinomycetes. Actinomycetes species have been mentioned in several reports as a source of bioactive secondary metabolites such as cytotoxic compounds (Yoo et al. 2002; Thangapandian et al. 2007) and antimicrobial compounds (Kumari et al. 2006; Rizk et al. 2007) that have the potential to control a wide range of pathogens. Millions of microorganisms, including indigenous actinomycetes, live in the marine environment and play an important role in the mineralization of complex organic matter, degradation of dead plants, plankton, and animals, removal of pollutants and toxicants, and production of primary and secondary metabolites (Genilloud et al. 2011). There are many unique and different types of actinomycetes in the maritime environment (Lam 2007). The isolation of novel secondary metabolites from distinct populations of actinomycetes from the marine environment suggests that these microorganisms represent an important new resource for marine and microbial natural product research (Abdel-Mageed et al. 2010; Asolkar et al. 2010; Li et al. 2011; Roh et al. 2011; Sousa et al. 2012; Jensen et al. 2005). Continuing attempts to study the variety of marine actinomycetes and how their development in the marine environment has influenced the generation of bioactive secondary metabolites have provided insights into how these microorganisms might be exploited to manufacture bioactive chemicals (Zhang et al. 2011).
Identification of the most potent strain AW6
Phenotypic identification
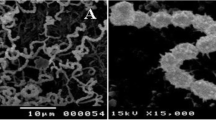
The morphological studies of the AW6 strain showed that the spore chain is rectiflexibiles with a smooth spore surface (Supplementary 1). The cultural properties of the AW6 strain after cultivation on different ISP media were summarized in Supplementary (2). Additionally, the physiological and biochemical characteristics for the selected isolate AW6 include nitrate reduction, catalase, methyl red (MR), indole production, Voges–Proskauer (V–P), starch hydrolysis, hydrogen sulfide production, and gelatin hydrolysis were also assessed and represented in Supplementary (3). The production of acid from carbohydrates was investigated with Actinomyces fermentation broth (BBL) as the basal medium. xylose, Arabinose, mannose, glucose, lactose, cellobiose, glycerol, raffinose, glycogen, salicin, mannitol, rhamnose, starch, inositol, and sorbitol were tested. The used tests in the biochemical study are performed as described in the report of the International Subgroup on the Taxonomy of Microaerophilic Actinomycetes (Slack 1968).
Genotypic identification
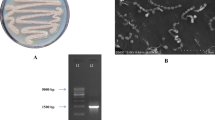
The 16S rRNA gene was extracted, amplified, sequenced, and aligned against known sequences deposited in the GeneBank database via the Basic Local Alignment Search Tool (BLAST) tool to measure the similarity score and calculate the statistical significance of the matches (http://www.blast.ncbi.nlm.nih.gov/Blast). The obtained result showed a very close similarity of the obtained sequence with 100% homology of the isolate AW6 with Actinomyces sp. Based on the analysis of the DNA sequence and the morphological characteristics of the AW6 isolate was identified as Actinomyces sp. AW6 and deposited in GenBank with accession no. OK090864.1. The maximum likelihood approach and the Tamura–Nei model were used to infer the evolutionary history (Fig. 1). The graph displays the tree with the highest log likelihood (− 17,134.78). The proportion of trees with the relevant taxa grouped is presented beside the branches. The tree is shown to scale, and branch lengths are quantified in terms of the number of substitutions per site. This study included 14 nucleotide sequences. The final dataset had a total of 1531 locations. MEGA-X was used to perform evolutionary analysis.
Large-scale production and purification of bioactive compounds
The strain Actinomyces sp. AW6 was cultivated on rice medium and extracted with ethyl acetate. The ethyl acetate evaporated using a rotary evaporator till complete evaporation of ethyl acetate. The extract (13 g) was primarily fractionated using Flash column chromatography into 100 fractions. Based on the chemical screening using TLC and biological screening, the most active fractions were collected together further and purified using Sephadex LH-20 column with a gradient mobile phase DCM: Methanol. The TLC chromatogram showed that fraction no. (8) contains the peaks of interest. The structural elucidation of the isolated compounds was performed using nuclear magnetic resonance (NMR) spectroscopy.
Compound (1) was obtained as white needles. 1H NMR spectra showed characteristic signals for ABX system of three aromatic protons at δH (ppm): 6.72 (IH, dd, J = 8.0, 2.1 Hz, H-6), 7.28 (1H, d, J = 8.0 Hz, H-5), and 6.75 (lH, d, J = 2.1 Hz, H-8) indicating to the presence of 1,2,4-trisubstituted benzene ring. Moreover, 1,2-disubstituted olefinic protons were resonated at δH (ppm): 6.19 (1H, d, J = 9.5 Hz, H-3), and 7.61 (1H, d, J = 9.5 Hz, H-4) indicating the presence of α,β-unsaturated ketone of a coumarin ring. 13C NMR spectra revealed the presence of characteristic signals for aromatic, olefinic and carbonyl carbons at δC (ppm): 162.0 (C-2), 113.7 (C-3), 145.0 (C-4), 129.89 (C-5), 112.1 (C-6), 160.97 (C-7), 102.94 (C-8), 156.2 (C-9), and 111.69 (C-10). Therefore, the compound could be identified as umbelliferone based on its chromatographic properties, proton and carbon spectra, and available reported data (Fig. 2) (Wagh et al. 2021).
Compound (2) was obtained as a pale orange powder. 1H NMR spectra showed characteristic signals for aromatic protons were resonated at δH (ppm): 7.10 (1H, s, H-2), 7.34 (1H, s, H-4), 7.33 (1H, d, J = 6.8, H-5), 7.74 (1H, dd, J = 8.2 and 6.8, H-6), and 7.51 (1H, d, J = 8.2, H-7) indicating the presence of quinone nucleus. Additionally, methyl and methoxy groups were resonated at δH (ppm): 2.38 (3H, s, –CH3) and 3.71 (3H, s, –OCH3). 13C NMR spectra revealed the presence of characteristic signals for aromatic, carbonyls, aromatic methyl, and methoxy carbons at δC (ppm): 157.0 (C-1), 115.7 (C-2), 118.9 (C-3), 122.36 (C-4), 118.7 (C-5), 137.1 (C-6), 122.9 (C-7), 160.93 (C-8), 185.0 (C-9), 182.5 (C-10), 132.6 (C-11), 113.9 (C-12), 138.9 (C-13), 128.2 (C-14), 56.1 (–OCH3-1), and 21.2 (–CH3-3) supporting the presence of anthraquinone skeleton. Based on its chromatographic properties, proton and carbon spectra, and available reported data, the compound could be identified as 1-methoxy-3-methyl-8-hydroxy-anthraquinone (Fig. 2) (Kumar et al. 2017).
Biological evaluation
Antimicrobial activity
The antimicrobial activity testing of compounds C1 and C2 using MTP assay revealed that compounds C1 and C2 have low antibacterial activity toward S. aureus and E. coli. While no pronounced activity has been detected toward the rest microbes including P. aeruginosa, C. albicans and A. niger. The minimum inhibitory concentration of the two compounds has been recorded in Table 3.
Antioxidant activity
The antioxidant activity of compounds (C1 and C2) based on 2,2-diphenyl-1-picrylhydrazyl radical (DPPH assay; 200 µg/mL) revealed that 1-methoxy-3-methyl-8-hydroxy-anthraquinone (C2) is the most antioxidant agent, showed maximum DPPH scavenging activity (55.25%), followed by umbelliferone)C1) (30.20%). Also, the two compounds showed DPPH antioxidant activity with IC50 values of 5.47 and 3.84 µg/mL for C1 and C2, respectively (Table 4).
In vitro DNA gyrase-B inhibition
DNA gyrase is a bacterial enzyme that catalyzes the ATP-dependent negative supercoiling of closed-circular double-stranded DNA. Gyrase is a member of the topoisomerase enzyme family, which is involved in the control of DNA topological transitions (Reece and Maxwell 1991). DNA gyrase is made up of two subunits: A and B. (Gyr-A and -B, respectively). Gyr-A is the component responsible for binding to DNA and relaxing its positive supercoils. Fluoroquinolone medicines are also effective against it. Gyr-B, on the other hand, is in charge of providing the necessary energy for this action by hydrolyzing one molecule of ATP (Zhuo et al. 2013). Accordingly, we tested the obtained purified compounds C1 and C2 for their Gyr-B inhibitory activity. As shown in Fig. 3, the two compounds showed inhibition of Gyr-B enzyme. Compound C1 was the most potent inhibitor, with an IC50 value of (3.79 ± 0.21 µM), while compound C2 was the least potent (IC50 = 13 ± 0.71 µM).
The ADME-related physicochemical properties
The ADME-related physicochemical properties of umbelliferone) C1) and 1-methoxy-3-methyl-8-hydroxy-anthraquinone (C2) ester were estimated using SwissADME online server (Daina et al. 2017). The measurements are based on the characterization of the purified compounds following the drug-likeness rules. Accordingly, the compound passed the Lipinski, Veber rules without any violation, while one violation (atoms < 20) has been detected with Ghose rule. On the other hand, (C2) passed the Lipinski, Veber, and Ghose rules filter with no violations. Both compounds exhibited 0.55% oral bioavailability and could be used as an oral medication (Table 5). Additionally, the rapid estimation of drug-likeness was performed by plotting of Bioavailability Radar plot of the two compounds which is based on six physicochemical parameters size, polarity, lipophilicity, solubility, flexibility, and saturation, while the pink area depicts the optimal value range for each parameter. According to the obtained diagram, the two compounds displayed optimal range (pink area) for all the parameters instauration parameter for C1 and C2 (Fig. 4a and b). Another important physicochemical parameter is Lipophilicity which shows the compound permeability across the cell membrane (Potts and Guy 1992; Rutkowska et al. 2013). Both tested compounds (C1 and C2) showed log Po/w values below 5, (1.50 and 2.48) suggesting a good permeability and absorption across the cell membrane. Additionally, solubility is one of the most important parameters influencing compound absorption in any formulation process (Daina et al. 2017). Based on ESOL topological model, compound 1 is soluble, while compound C2 is moderately soluble. For defining medicinal chemistry and Leadlikness, compound C1 did not pass the rule of three (RO3), as it has one violation for this rule, while compound 2 passed the rule of three (RO3), with no violation. For synthetic accessibility score (SAscore) that estimated on the similarity of fragments and complexity penalties, both compounds (C1 and C2) showed moderate synthetic accessibility with values (2.56 and 2.57), respectively.
Bioavailability Radar plot of a compound 1 and b compound 2. The pink area shows the optimal range for each property (Lipophilicity: XLOGP3 between − 0.7 and + 5.0, size: MW between 150 and 500 g/mol, polarity: TPSA between 20 and 130 Å2, solubility: log S not higher than 6, saturation: fraction of carbons in the sp3 hybridization not less than 0.25, and flexibility: no more than 9 rotatable bonds) (color figure online)
Table 5, showed the compounds (C1 and C2) pharmacokinetic parameters measured using the vector machine algorithm (SVM) model (Daina et al. 2017). Compound C1 showed selective inhibitory activity toward CYP1A2 isoenzyme, while compound C2 showed selective activity against CYP1A2, CYP2C19, and CYP2C9 isoenzymes. Figure 5 represents the BOILED-Egg model (Brain or Intestinal Estimate D permeation method, WLOGP vs TPSA) that was adapted from Daina et al (2017). The compounds (C1 and C2) demonstrated high human gastrointestinal absorption (GI). The compounds are non-P-gp substrates (PGP-, red dots), while their blood–brain barrier (BBB) permeant (TPSA < 75 Å2), suggesting the presence of effects on the central nervous system (CNS) (Daina and Zoete 2016). Prediction of skin permeability coefficient (Kp) of the obtained compound was done as described by Potts and Guy (Potts and Guy 1992). The two compounds (C1 and C2) showed log (Kp) (− 6.17 and − 5.52 cm/s), while the more negative log Kp, the less skin permeant is the compounds.
BOILED-Egg plot for both C1 and C2. The yellow zone (yolk) is for highly possible BBB permeability, while the white region (GI) is for highly probable HIA (GI) absorption. Molecules with minimal absorption and no brain penetration are represented by the outside gray zone. The points are also colored blue if P-gp substrate (PGP +) is expected, and red if P-gp non-substrate (PGP) is projected (color figure online)
Toxicity prediction via ProTox ii
Toxicity prediction of the obtained compounds (C1 and C2) was performed using ProTox ii webserver (Banerjee et al. 2018). Results in the Table 6 showed that compound 1 has no pronounced toxicity, while compound 2 showed activity toward some targets as predicted by ProTox ii.
Conclusion
Actinobacteria are the most widely distributed microorganisms on earth with the ability to produce unique bioactive compounds with high commercial value. The strain Actinomyces sp. AW6 showed pronounced antioxidant and antimicrobial potential against E. coli, S. aureus, B. subtilis, P. aeruginosa, A. niger and C. albicans. Cultivation of the selected strain led to the isolation of two compounds C1: umbelliferone and C2: 1-methoxy-3-methyl-8-hydroxy-anthraquinone. Both compounds displayed antibacterial activity toward S. aureus and E. coli. They also exhibited antioxidant activity and anti-Gyr-B enzyme activity with IC50 value of (3.79 ± 0.21 µM) for C1, and (IC50 = 13 ± 0.71 µM) for C2. The ADME-related physicochemical properties of the obtained compound were predicted using SwissADME web tools and the ProToxii webserver was used to estimate in silico toxicity.
Code availability
Not applicable.
References
Abdel-Aziz MS, Ghareeb MA, Saad AM, Refahy LA, Hamed AA (2018) Chromatographic isolation and structural elucidation of secondary metabolites from the soil-inhabiting fungus Aspergillus fumigatus 3T-EGY. Acta Chromatogr 30(4):243–249. https://doi.org/10.1556/1326.2017.00329
Abdel-Mageed WM, Milne BF, Wagner M, Schumacher M, Sandor P, Pathom-aree W, Goodfellow M, Bull AT, Horikoshi K, Ebel R, Diederich M, Fiedlerd H, Jaspars M (2010) Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Org Biomol Chem 8:2352–2362. https://doi.org/10.1039/C001445A
Alhadrami HA, Orfali R, Hamed AA, Ghoneim MM, Hassan HM, Hassane ASI, Rateb ME, Sayed AM, Gamaleldin NM (2021) Flavonoid-coated gold nanoparticles as efficient antibiotics against gram-negative bacteria—evidence from in silico-supported in vitro studies. Antibiotics 10:968. https://doi.org/10.3390/antibiotics10080968
Asolkar RN, Kirkland TN, Jensen PR, Fenical W (2010) Arenimycin, an antibiotic effective against rifampin- and ethicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J Antibiot 63(1):37–39. https://doi.org/10.1038/ja.2009.114
Banerjee P, Eckert AO, Schrey AK, Preissner R (2018) ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 2(46):W257–W263. https://doi.org/10.1093/nar/gky318
Bergey DH (1989) Bergey`s Manual of Systematic Bacteriology. vol 4. Williams and Wilkins Co., Baltimore, ISBN: 0-683-09061-5
Brand-Williams W, Cuvelier ME, Berset C (1995) Use of a free-radical method to evaluate antioxidant activity. LWT Food Sci Technol 28(1):25–30. https://doi.org/10.1016/S0023-6438(95)80008-5
Burhamzah R, Djide MN, Rante H, Zainuddi EN (2016) Isolation and screening of antimicrobial-producing actinomycetes from marine sediment of Galesong Coast, Indonesia. Asian J Microbiol Biotechnol Environ Sci 18(1):31–34
Daina A, Zoete V (2016) A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. Chem Med Chem 11:1117–1121. https://doi.org/10.1002/cmdc.201600182
Daina A, Michielin O, Zoete V (2017) Swiss ADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717–42730. https://doi.org/10.1038/srep42717
Durcik M, Tammela P, Barančoková M, Tomašič T, Ilaš J, Kikelj D, Zidar N (2018) Synthesis and evaluation of N-phenylpyrrolamides as DNA gyrase B Inhibitors. Chem Med Chem 13:186–198. https://doi.org/10.1002/cmdc.201700549
Elkhouly HI, Hamed AA, El Hosainy AM, Ghareeb MA, Sidkey NM (2021a) Bioactive secondary metabolite from endophytic Aspergillus Tubenginses ASH4 isolated from Hyoscyamus muticus: Antimicrobial, antibiofilm, antioxidant and anticancer activity. Pharmacogn J 13(2):434–442. https://doi.org/10.5530/pj.2021.13.55
Elkhouly HI, Sidkey NM, Ghareeb MA, El Hosainy AM, Hamed AA (2021b) Bioactive secondary metabolites from endophytic Aspergillus terreus AH1 isolated from Ipomoea carnea growing in Egypt. Egypt J Chem 64(12):7511–7520. https://doi.org/10.21608/EJCHEM.2021.85908.4161
El-Shazly MAM, Hamed AA, Kabary HA, Ghareeb MA (2021) LC-MS/MS profiling, antibiofilm, antimicrobial and bacterial growth kinetic studies of Pluchea dioscoridis extracts. Acta Chromatogr. https://doi.org/10.1556/1326.2021.00956
Gardete S, Tomasz A (2014) Mechanisms of vancomycin resistance in Staphylococcus aureus. J Clin Invest 124(7):2836–2840. https://doi.org/10.1172/JCI68834
Genilloud O, González I, Salazar O, Martín J, Tormo JR, Vicente F (2011) Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol 38:375–389. https://doi.org/10.1007/s10295-010-0882-7
Ghareeb MA, Refahy LA, Saad AM, Osman NS, Abdel-Aziz MS, El-Shazly MA, Mohamed AS (2015) In vitro antimicrobial activity of five Egyptian plant species. J Appl Pharma Sci 5(2):045–049. https://doi.org/10.7324/JAPS.2015.58.S7
Hamed AA, Kabary H, Khedr M, Emam AN (2020a) Antibiofilm, antimicrobial and cytotoxic activity of extracellular green-synthesized silver nanoparticles by two marine-derived actinomycete. RSC Adv 10:10361–10367
Hamed AA, Soldatou S, Qader MM, Arjunan S, Miranda KJ, Casolari F, Pavesi C, Diyaolu OA, Thissera B, Eshelli M, Belbahri L, Luptakova L, Ibrahim NA, Abdel-Aziz MS, Eid BM, Ghareeb MA, Rateb ME, Ebel R (2020b) Screening fungal endophytes derived from under-explored Egyptian marine habitats for antimicrobial and antioxidant properties in factionalised textiles. Microorganisms 8:1617. https://doi.org/10.3390/microorganisms8101617
Hamed AA, Eskander DM, Badawy MSEM (2022) Isolation of secondary metabolites from marine Streptomyces sparsus ASD203 and evaluation its bioactivity. Egypt J Chem 65:539–547
Jensen PR, Mincer TJ, Williams PG, Fenical W (2005) Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87:43–48. https://doi.org/10.1007/s10482-004-6540-1
Kest H, Kaushik A (2019) Vancomycin-Resistant Staphylococcus aureus: formidable threat or silence before the storm? J Infect Dis Epidemiol 5(5):93. https://doi.org/10.23937/2474-3658/1510093
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Kumar CG, Mongolla P, Chandrasekhar C, Poornachandra Y, Siva B, Babu KS, Ramakrishna KVS (2017) Anti-proliferative and antioxidant activities of 1- methoxy-3-methyl-8-hydroxy-anthraquinone, a hydroxyanthraquinoid extrolite produced by Amycolatopsis thermoflava strain SFMA-103. Microbiol Biotechnol Lett 45(3):200–208. https://doi.org/10.4014/mbl.1705.05001
Kumari KK, Ponmurugan P, Kannan N (2006) Isolation and characterization of Streptomyces sp. for secondary metabolite production. Biotechnology 5(4):478–480. https://doi.org/10.3923/biotech.2006.478.480
Lam KS (2007) New aspects of natural products in drug discovery. Trends Microbiol 15(6):279–289. https://doi.org/10.1016/j.tim.2007.04.001
Li S, Tian X, Niu S, Zhang W, Chen Y, Zhang C (2011) Pseudonocardians AC new diazaanthraquinone derivatives from a deap-sea actinomycete Pseudonocardia sp. SCSIO 01299. Mar Drugs 9(8):1428–1439. https://doi.org/10.3390/md9081428
Mohammed HS, Abdel-Aziz MM, Abu-baker MS, Saad AM, Mohamed MA, Ghareeb MA (2019) Antibacterial and potential antidiabetic activities of flavone C-glycosides isolated from Beta vulgaris subspecies cicla L. var. flavescens (Amaranthaceae) cultivated in Egypt. Curr Pharm Biotechnol 20(7):595–604. https://doi.org/10.2174/1389201020666190613161212
Moradali MF, Ghods S, Rehm BHA (2017) Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 7:39. https://doi.org/10.3389/fcimb.2017.00039
Palazzotto E, Tong Y, Lee SY, Weber T (2019) Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol Adv 37(6):107366. https://doi.org/10.1016/j.biotechadv.2019.03.005
Potts RO, Guy RH (1992) Predicting skin permeability. Pharm Res 9:663–669. https://doi.org/10.1023/A:1015810312465
Pradhan P, Tamang JP (2019) Phenotypic and genotypic identification of bacteria isolated from traditionally prepared dry starters of the Eastern Himalayas. Front Microbiol 10:2526. https://doi.org/10.3389/fmicb.2019.02526
Qader MM, Hamed AA, Soldatou S, Abdelraof M, Elawady ME, Hassane ASI, Belbahri L, Ebel R, Rateb ME (2021) Antimicrobial and antibiofilm activities of the fungal metabolites isolated from the marine endophytes Epicoccum nigrum M13 and Alternaria alternata 13A. Mar Drugs 19(4):232. https://doi.org/10.3390/md19040232
Reece RJ, Maxwell A (1991) DNA gyrase: structure and function. Crit Rev Biochem Mol Biol 26(3–4):335–375. https://doi.org/10.3109/10409239109114072
Rizk M, Abdel-Rahman T, Metwally H (2007) Screening of antagonistic activity in different Streptomyces species against some pathogenic microorganisms. J Boil Sci 7(8):1418–1423. https://doi.org/10.3923/jbs.2007.1418.1423
Roh H, Uguru GC, Ko HJ, Kim BY, Goodfellow M, Bull AT, Stach JE (2011) Genome sequence of the abyssomicin- and proximicin-producing marine actinomycete Verrucosispora maris AB-18-032. J Bacteriol 193(13):3391–3391. https://doi.org/10.1128/JB.05041-11
Rutkowska E, Pajak K, Jozwiak K (2013) Lipophilicity-methods of determination and its role in medicinal chemistry. Acta Pol Pharm 70(1):3–18
Singh S, Rawat A, Sharma DC (2012) Antifungal antibiotic production by Streptomyces sp. isolated from soil. Int J Cur Res Rev 4:7–16
Slack JM (1968) Subgroup on the taxonomy of Microaerophilic actinomycetes. Report on organization, aims and procedures. Intern J Syst Bacteriol 18:253–262. https://doi.org/10.1099/00207713-18-3-253
Sousa TS, Jimenez PC, Ferreira EG, Silveira ER, Braz-Filho R, Pessoa ODL, Costa-Lotufo LV (2012) Anthracyclinones from Micromonospora sp. J Nat Prod 75(3):489–493. https://doi.org/10.1021/np200795p
Thangapandian V, Ponmurugan P, Ponmurugan K (2007) Actinomycetes diversity in the rhizosphere soils of different medicinal plants in Kolly Hills-Tamilnadu, India, for secondary metabolite production. Asian J Plant Sci 6(1):66–70. https://doi.org/10.3923/ajps.2007.66.70
Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VGJ (2015) Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28(3):603–661. https://doi.org/10.1128/CMR.00134-14
van der Meij A, Worsley SF, Hutchings MI, van Wezel GP (2017) Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev 41(3):392–416. https://doi.org/10.1093/femsre/fux005
Ventola CL (2015) The antibiotic resistance crisis part 1: causes and threats. Pharm Ther 40(4):277–283
Wagh A, Butle S, Raut D (2021) Isolation, identification, and cytotoxicity evaluation of phytochemicals from chloroform extract of Spathodea campanulata. Futur J Pharm Sci 7(58):1–8. https://doi.org/10.1186/s43094-021-00205-7
Waithaka P, Mwaura FB, Wagacha M, Gathuru E, Githaiga BM (2017) Antimicrobial properties of actinomycetes isolated from Menengai Crater in Kenya. Cell Bio 6:13–26. https://doi.org/10.4236/cellbio.2017.62002
Yoo JC, Han JM, Nam SK, Baik KS, Jo JS, Seong CN (2002) Characterization of a Streptomycete isolate producing the potent cytotoxic substance, nonadecanoic acid. J Microbiol 40(2):178–181
Zhang DD, Lee HF, Wang C, Li B, Zhang J, Pei Q, Chen J (2011) Climate change and large scale human population collapses in the pre-industrial era. Glob Ecol Biogeogr 20:520–531. https://doi.org/10.1111/j.1466-8238.2010.00625.x
Zhuo S-T, Li C-Y, Hu M-H, Chen S-B, Yao P-F, Huang S-L, Ou T-M, Tan J-H, An L-K, Li D (2013) Synthesis and biological evaluation of benzo [a] phenazine derivatives as a dual inhibitor of topoisomerase I and II. Org Biomol Chem 11:3989–4005. https://doi.org/10.1039/c3ob40325d
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). The authors would like to thank the Ain Shams University and National Research Centre, Egypt for the support and funding.
Author information
Authors and Affiliations
Contributions
MAA: performed the experiments. MAA, MAG, AAH and MKI: wrote the manuscript, analyzed the data. MAG, EAA, AAH, and MKI: guided the experiments and all authors revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Agour, M.A., Hamed, A.A., Ghareeb, M.A. et al. Bioactive secondary metabolites from marine Actinomyces sp. AW6 with an evaluation of ADME-related physicochemical properties. Arch Microbiol 204, 537 (2022). https://doi.org/10.1007/s00203-022-03092-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03092-5