Abstract
The bacterial foodborne pathogen Listeria monocytogenes has been implicated in fresh produce outbreaks with a significant economic impact. Given that L. monocytogenes is widespread in the environment, food production facilities constantly monitor for the presence of Listeria species. To develop a surveillance platform for food processing facilities, this study conducted a comparative genomic analysis for the identification of conserved high copy sequences in the ribosomal RNA of Listeria species. Simulated folding was performed to assess RNA accessibility in the identified genomic regions targeted for detection, and the developed singleplex assay accurately detected cell amounts lower than 5 cells, while no signals were detected for non-targeted bacteria. The singleplex assay was subsequently tested with a flow-through system, consisting of a DNA aptamer-capture step, followed by sample concentration and mechanical lysis for the detection of Listeria species. Validation experiments indicated the continuous flow-through system accurately detected Listeria species at low cell concentrations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foodborne pathogens are responsible for a consistent level of human illness that poses a substantial public health and economic burden, resulting in an economic burden of $15.5 billion annually (Hoffmann et al. 2015). Listeriosis, a serious infection caused by eating food contaminated with the bacterium Listeria monocytogenes, has been recognized as an important public health problem in the United States, and the annual economic impact of listeriosis in the United States alone is estimated at over US$2.8 billion. There has been an estimated 1500 listeriosis cases each year, and of those, approximately 300 cases have resulted in death (Hoffmann et al. 2012; Scallan et al. 2011). In particular, L. monocytogenes is responsible for systemic listeriosis with an approximate 30% mortality rate in susceptible populations of pregnant women, neonates, elderly, or immunocompromised patients (Buchanan et al. 2017; Hoffmann and Scallan Walter 2019; Scallan et al. 2011).
While L. monocytogenes is considered one of the major foodborne pathogens that cause listeriosis in humans and animals (Pesavento et al. 2010), other Listeria species, which include Listeria grayi, Listeria innocua, Listeria ivanovii, Listeria seeligeri, and Listeria welshimeri, have been occasionally implicated in human clinical case reports, primarily in individuals with suppressed immune functions and/or underlying illnesses (Korsak and Szuplewska 2016). Listeria species are widely distributed in various environments, including soil, water, vegetation, animal feed, farm environments, food processing environments, urban and suburban environments (Korsak and Szuplewska 2016). L. monocytogenes and L. innocua have been the most prevalent species of Listeria found in urban environments, produce production, preharvest environments, retail environments, and processing environments (Estrada et al. 2020). Recently, high profile outbreaks of L. monocytogenes have been associated with deli meats, fresh produce, and ready-to-eat foods (Churchill et al. 2019). Other Listeria species have also been found in ready-to-eat, raw or unprocessed foods (Arslan and Özdemir 2020; Guerra et al. 2001; Pesavento et al. 2010; Soriano et al. 2001; Zeinali et al. 2017), as well as in food processors (Huang et al. 2007; Korsak and Szuplewska 2016) and other food products (Chen et al. 2009; Pesavento et al. 2010). Given that L. monocytogenes is widespread in the environment, food production facilities constantly monitor and control for the presence of Listeria species on surfaces. Listeria species are considered a broad indicator of the conditions potentially favorable for L. monocytogenes growth and survival in the environment (Brouillette et al. 2014; The United Fresh Food Safety & Technology Council 2018; Zoellner et al. 2018). Using a broad indicator group, such as screening for all Listeria species (genus level), increases the chances of finding L. monocytogenes niches and reacting in an effective manner to mitigate the prevalence of this pathogen in a food production facility (Brouillette et al. 2014; The United Fresh Food Safety & Technology Council 2018; Zoellner et al. 2018).
Given that conventional culture-based assays are labor intensive and time consuming, several methodologies have been developed for the identification of foodborne pathogens, which can be further classified into nucleic acid-based, biosensor-based and immunological-based methods (Chen et al. 2017; Law et al. 2015). In particular, nucleic acid-based methods, employing real-time quantitative PCR (qPCR), have become preferred methods for the detection and quantification of Listeria species due to their simplicity, high sensitivity and specificity, and low risk of contamination due to the lack of post-processing steps for obtaining the detectable signal when compared to conventional PCR (Chen et al. 2017; Gasanov et al. 2005). Recently, several multiplex qPCR assays have become available for the identification of multiple Listeria species by targeting genes required for virulence and regulatory functions including those coding for bacteriolytic properties (iap), phosphatidylinositol phospholipase C (plcB), sucrose-specific enzyme (scrA) as well as putative internalin and oxidoreductase, and N-acetylmuramidase proteins (Chen et al. 2017; Hage et al. 2014; Hein et al. 2001; Hitchins et al. 2017). Other qPCR assays have been developed by targeting the 23S ribosomal DNA or non-coding RNA specific to the Listeria genus; however, these qPCR assays require multiple sets of primers and probes to be multiplexed to enable the detection of various Listeria species (Chen et al. 2017; Petrauskene et al. 2017; Rodríguez-Lázaro et al. 2004).
In the past years, there has been a significant increase in the availability of bacterial genome sequences in public databases (Land et al. 2015; Sayers et al. 2020). These recent developments would consequently enable the improvement of the new design of singleplex qPCR assays for detecting multiple strains of Listeria species by allowing better identification of specific and accessible genomic regions to be inclusive in detecting the targeted species while still being exclusive in discriminating against non-Listeria strains. Given that ribosomal RNA (rRNA) is present in large quantities and multiple copies in bacteria, in particular in the Listeria genome (Glaser et al. 2001; Milner et al. 2001), new approaches on the development of molecular assays for detecting foodborne pathogens have chosen RNA as the targeted analyte (Livezey et al. 2013), which offers a reliable and highly sensitive detection of Listeria species. In the present study, a comparative genomic analysis was conducted for evaluating high copy-number sequences to enable the targeted and specific detection of Listeria species. By conducting an in silico analysis with a dynamic programming algorithm, simulated folding was performed to assess the accessibility of multicopy targeted regions in the rRNA for optimal detection. This nucleic acid-based assay, a singleplex assay, was further validated by conducting inclusivity tests for Listeria species and exclusivity tests for non-targeted environmental bacterial strains, belonging to the Bacillus, Citrobacter, Enterobacter, Pseudomonas, Salmonella and Shigella genera. As a proof-of-concept for the development of an in-process detection system for foodborne pathogens in food processing facilities, this nucleic acid-based assay was tested in conjunction with a flow-through system, consisting of aptamer-capture step, followed by sample concentration and mechanical lysis for the detection of Listeria species at low cell concentrations.
Materials and methods
Bacterial strains and culture conditions
The Listeria strains used in the present study (Table 1) were streaked for isolation on BBL™ Trypticase™ Soy Agar (Becton, Dickinson and Co., Franklin Lakes, NJ) with 0.6% Bacto™ Yeast Extract (Becton, Dickinson and Co.) (Hitchins et al. 2017). Listeria species liquid cultures were grown in BD™ Tryptic Soy Broth (Becton, Dickinson and Co., Franklin Lakes, NJ) or Brain Heart Infusion (BHI) Broth (K25; Hardy Diagnostics, Santa Maria, CA) and were further incubated overnight with constant shaking (225 rpm) at 37 °C. Non-targeted bacterial strains (Table 1) were streaked for isolation on Luria–Bertani agar (LB; Becton, Dickinson, and Co.), and liquid cultures were grown in LB broth overnight with constant shaking (225 rpm) incubated at 37 °C for strains belonging to the genera Citrobacter, Escherichia, Salmonella, and Shigella and incubated at 28 °C for strains belonging the genera Bacillus, Enterobacter, and Pseudomonas.
Comparative genomics and in silico analysis for oligonucleotide design
To design oligonucleotides targeting the rRNA operon (rrn operon), 59 ribosome sequences of 19 Listeria species including the common Listeria species: L. innocua, L. ivanovii, L. grayi, L. monocytogenes, and L. welshimeri. L. monocytogenes strains with all known serovars were included for the design (Doumith et al. 2004). The ribosome sequence of L. monocytogenes strain EGD-e (GenBank Accession No. CP023861) was chosen as a model ribosome sequence (Glaser et al. 2001). Non-targeted bacterial sequences were selected by searching the top non-Listeria matches with the model ribosome sequence. Additional bacterial strains belonging to a diverse genera such as Citrobacter, Enterobacter, and Pseudomonas, commonly found as resident bacteria in agricultural and food processing environments (Dees et al. 2015; Møretrø and Langsrud 2017; Orsi and Wiedmann 2016; Williams and Marco 2014; Williams et al. 2013), were also added to the non-targeted list of genomes examined. Simulated folding of the model sequence was performed to find regions of high RNA accessibility with unfolding energy < 20 kcal/mol using Visual-OMP™ software package (DNA Software, Inc., Ann Arbor, MI). The target sequences and non-target sequences were aligned using Geneious Software (Biomatters, Ltd., Aukland, NewZealand). Regions of high RNA accessibility in the model ribosome sequence with the most heterogeneity with an average of less than 80% match with the reference sequence were selected as target regions for the primer design using RealTimeDesign™ qPCR Assay Design Software (LGC, Biosearch Technologies, Petaluma, CA). The use of the RealTimeDesign™ software allowed the design with the proprietary BHQplus® probes incorporating a duplex-stabilizing technology (LGC, Biosearch Technologies) to elevate the melting temperature and enhance target specificity (Kutyavin 2008). The default of least restrictive parameters for RealTimeDesign™ software were used in the design (Sowers et al. 2005, 2006); however, for some regions, the default parameters were modified to design oligonucleotides with increased specificity to the model target. In particular, the minimum primer temperature was lowered from 65 °C to 60 °C, and the maximum probe temperature was increased from 72 °C to 77 °C. Additionally, the maximum internal stability was increased to 0.6 from 0.0. In some instances, runs of four guanidine bases were allowed to improve specificity. Three guanidine and/or cytosine bases were allowed at the 3’ end of the oligonucleotide, and the maximum percentage of the guanidine bases in the oligonucleotide probes was increased to 40%.
Nucleic acid extractions
Purified RNA was extracted from diluted overnight broth cultures using the ClaremontBio RNAexpress with OmniLyse® HL Kit and DNAexpress columns (Claremont BioSolutions, LLC, Upland, CA) or the Qiagen RNeasy® Protect Bacteria Mini Kit (QIAGEN, Inc., Valencia, CA), according to the manufacturer’s guidelines. The quantity of nucleic acid was assessed by fluorometric measurement using the Qubit™ 4 Fluorometer (Invitrogen, Carlsbad, CA), and the quality was evaluated using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Crude lysates and plate enumerations were prepared by aliquoting diluted overnight broth culture at mid-log (OD600 absorbance ~ 0.2–0.3) and serially diluting tenfold in nuclease-free water and 1 × phosphate-buffered saline (PBS), respectively. To prepare crude lysates, each serially diluted culture in nuclease-free water was lysed by mechanical disruption using an OmniLyser (Claremont BioSolutions, LLC). For plate enumerations, a 10 µl droplet of each serially diluted culture in 1 × PBS was plated on fresh solid LB agar overnight at 37 °C for Citrobacter species, Escherichia species, Listeria species, Salmonella species, and Shigella species strains and 28 °C for Bacillus species, Enterobacter species, and Pseudomonas species.
Nucleic acid amplifications
Using a unidirectional workflow previously employed for experimental procedures with foodborne enteric viruses (Quiñones et al. 2017), the amplification by reverse transcription-quantitative PCR (RT-qPCR) was performed in a 20 µl reaction mixture containing 5 µl of 4 × TaqPath™ 1-Step Multiplex Master Mix (Applied Biosystems, Foster City, CA), 0.5 µM of each forward and reverse BH1 primer, 0.1 µM of BH1 probe (LGC, Biosearch Technologies) and 1 µl purified RNA or 5 µl crude lysate. The reaction mixtures were placed in a QuantStudio™ 5 Real-Time PCR System (Applied Biosystems) with the following parameters: RT step at 54 °C for 15 min, hot start at 95 °C for 2 min followed by 40 cycles each of denaturation at 95 °C for 3 s and annealing and extension at 64 °C for 30 s. Cycle threshold (Ct) values were analyzed using the QuantStudio™ Design & Analysis software (Applied Biosystems). The performance of the BH1 oligonucleotide-based assay was compared with a DNA-based assay, MicroSEQ™ Listeria monocytogenes Detection kit (Applied Biosystems) with tenfold serial dilutions of L. monocytogenes cell lysate. The estimated cell concentrations in the suspension were determined by optical density and confirmed by viable bacterial colony count on solid media.
Design of a continuous flow-through detection system
For the design and validation of the continuous flow-through detection system, L. grayi strain RM2208 was used as the model organism (Table 1). In detail, samples were blended with capture buffer (0.2 × PBS, 2 mM magnesium chloride, and 0.2% Triton X-100) and filtered into a sample container using a Whirl–Pak® sterile sampling bag with 330 µm filter membrane (B01547; Nasco, Fort Atkinson, WI) to remove particulates. A biocompatible Masterflex™ peristaltic pump (Masterflex™ Model 7550–60; Cole-Parmer Instrument Co., Vernon Hills, IL) introduced the liquid sample into a Tenny Jr. temperature-controlled chamber (Model TLJR; Tenney Environmental, Parsippany, NJ) (Medin et al. 2020). Fluidic valves (Model SMC LVM205R-6B1; SMC Pneumatics, Yorba Linda, CA) moved homogenized sample through a depth filter, consisting of glass beads (100 µm diameter; BioSpec Products, Inc., Bartlesville, OK) having non-fouling surface properties (Medin et al. 2020). The Listeria cells were then captured in the column using aptamer-functionalized soda-lime glass beads (100 µm diameter; BioSpec Products, Inc.) with silane surface modification for attachment of the oligonucleotide sequences, serving as a spacer sequence (5′-GTTTTTGTTTTGAAAGTTGTTTTTTTTTT-3′) for aptamer extension and a tether sequence (5′-CAACTTTCAAAACAAAAACTTTTTTTTTT-Amino C6-3′) for aptamer surface attachment (Medin et al. 2020; Quiñones et al. 2020). The release of the sequestered Listeria cells from the column using release buffer (1 M NaCl, 100 mM EDTA, 0.1% Tween 20, 0.05% SDS, 30 mM NaHCO3, pH 10.3–10.4) (Medin et al. 2020). After potential inhibitors were removed as waste, the released Listeria cells were mechanically lysed using an ultrasonic transducer (Black & Decker, Baltimore, MD) and yttrium-stabilized zirconia beads (100 µm diameter; Pingxiang Chemshun Ceramics Co. Ltd., Pingxiang City, Jiangxi Province, China). An aliquot of the lysate was subjected to further amplification by RT-qPCR by using the BH1 oligonucleotide assay. To determine the capture efficiency of the aptamer-functionalized column, the capture column was conditioned by adding approximately 30 ml of capture buffer through the column at 15 ml/min using a Masterflex™ peristaltic pump (Cole-Parmer Instrument Co.), and the conditioning flow-through was discarded. A 10 ml Listeria cell suspension, diluted to a final concentration of approximately 1.6 × 105 CFU/ml, was added to the capture column. The entire flow-through was collected, and the L. grayi cell concentration was enumerated. The capture efficiency to the aptamer column was determined as the fraction of Listeria cells from the starting cell suspension added to the capture column that were not detected in the flow-through cell count, as previously described (Medin et al. 2020).
Statistical analysis
Statistical significance in the detected Ct values was determined by performing two-tailed Fisher’s exact test using R statistical software (version 3.0.1; R Foundation for Statistical Computing, Vienna, Austria) (Fisher 1935; Mehta and Patel 1986). Descriptive statistical analyses and one-way analysis of variance (ANOVA) were performed using the Analysis ToolPak in Microsoft® Excel® version 2002 for Office 365 ProPlus (Microsoft Corporation, Redmond, WA). P-values of ≤ 0.005 were considered statistically significant.
Results and discussion
Design strategy of the oligonucleotide-based assay for detecting Listeria species
As a suitable targeted region, rRNA has been considered for the development of detection assays as it is found to be represented in the bacterial genome in multiple copies (Livezey et al. 2013; Milner et al. 2001). In particular, whole-genome analysis revealed that a single L. monocytogenes cell contains 6 copies of the rRNA (rrn) operon (Glaser et al. 2001), and expression studies estimated that approximately 600–25,000 copies of the ribosomes were detected per cell (Milner et al. 2001). These findings have indicated that targeting rRNA is more than enough for a reliable amplification of nucleic acids to enable detection of the targeted pathogen at low cell concentrations. To initiate the design of oligonucleotides, approximately 200 genomic entries of distinct species of Listeria and non-targeted bacterial species were aligned and compared, and regions in the ribosome sequences with the most heterogeneity between the Listeria sequences and non-target sequences were selected for the probe design (Fig. 1). The distinct species of Listeria for the comparative genomic analysis included Listeria aquatica, Listeria booriae, Listeria cornellensis, Listeria costaricensis, Listeria goaensis, Listeria fleischmannii, Listeria floridensis, Listeria grandensis, L. grayi, L. innocua, L. ivanovii, Listeria marthii, L. monocytogenes, Listeria newyorkensis, Listeria riparia, Listeria rocourtiae, L. seeligeri, Listeria thailandensis, Listeria weihenstephanensis, and L. welshimeri.
Design strategy and specificity of the oligonucleotide-based assay for detecting Listeria species. Ribosomal RNA (rRNA) was chosen as the targeted region to enable a reliable detection of Listeria species at low cell concentrations. a Simulated folding was performed to assess RNA accessibility in the identified regions (circles) by calculating the equilibrium target unfolding (blue symbols) and the target complexity (green symbols) calculations, as determined using Visual-OMP™ software package (DNA Software, Inc., Ann Arbor, MI). b To further assess the oligonucleotide specificity for cross hybridization with non-targeted bacterial species from soil, water, and plant surfaces, the in silico mismatch was examined by comparing Listeria monocytogenes sequences with strains belonging to Bacillus, Citrobacter, Escherichia, Lactobacillus, Pantoea, Pectobacterium, Planococcus, Proteus, Pseudomonas, and Salmonella genera
The in silico analyses were performed with algorithms using the nearest-neighbor model coupled with multi-state equilibrium model for conducting simulated folding to assess RNA accessibility in the identified genomic regions targeted for detection (Fig. 1a). The results revealed multiple locations in the genome with an optimal target complexity as well as target unfolding. As shown in Fig. 1a, analysis of the 16S rRNA in L. monocytogenes revealed that the targeted region for the oligonucleotide design had an unfolding delta G calculation to be approximately 14 K to 16 K cal/mol when compared to most sites in the examined region with much higher values of 26–28 K cal/mol. To reduce any false positives due to low levels of specificity in the oligonucleotide design, the complexity of the targeted regions was also further examined (Fig. 1a). The analysis of optimal genome locations was further assessed to select the best sites for designing the targeted oligonucleotides (Fig. 1a, circles), and these selected sites for the design of the oligonucleotides were found to have optimal complexity scores between 0.8 and 1, indicating relative accessibility and complexity of regions within the target sequences (Nielsen et al. 2003). Additionally, the in silico mismatch was examined by comparing a collection of various Listeria species genome sequences with other bacterial non-targets, commonly present from soil, water, and plant surfaces (Fig. 1b). The sequence alignment to non-targeted bacterial species, included Bacillus species,Escherichia species, Pseudomonas species, and Citrobacter species, representing non-targeted bacterial species from agricultural environments (Dees et al. 2015; Orsi and Wiedmann 2016; Williams and Marco 2014; Williams et al. 2013).
To increase the level of desired target specificity at the genus level, the oligonucleotide design targeted the rrn operon comprising of the 16S and 23S rRNA gene clusters in the Listeria genome, a region commonly used as a microbial molecular marker for phylogenetic classification of bacterial species (Woese 1987). The design employed base analogs to promote nucleic acid duplex stabilizing, enabling an elevated melting temperature of the oligonucleotides (Kutyavin 2008). Additional standard criteria were also employed for developing the nucleic acid amplification assays, which consisted of examining for the G–C content, annealing temperatures, and self-hybridization (Sowers et al. 2005, 2006). The in silico analysis resulted in a total of six oligonucleotide sets with a G–C content ranging from 36.8% to 57.9% and annealing temperatures ranging from 55.4 °C to 76.6 °C (Table 2).
Assay specificity for target RNA detection in Listeria species
As a preliminary screening test, the various candidates of designed oligonucleotides sets were subjected to an initial inclusivity test using L. monocytogenes as a representative targeted species, and an exclusivity test was conducted using B. cereus as a representative non-targeted species, which was based on previous whole-genome comparisons suggesting that high conservation in the genome organization is specific for Bacillus and Listeria (Buchrieser et al. 2003). Amplifications for detecting L. monocytogenes with 10 fg of RNA as template, corresponding to 10 cell equivalents (Milner et al. 2001), resulted in Ct values, ranging from 28.5 to 33.7 for all of the oligonucleotide sets (Table 2). However, only the BH1 oligonucleotide set was found to show no cross reactivity for detecting B. cereus in the exclusivity test even at template amounts that were 100-fold when compared to the targeted Listeria species. The other candidate oligonucleotide sets showed a positive reaction resulting in Ct values ranging from 16.4 to 32.5 (Table 2). Based on these initial observations determining the specificity of the amplification reaction, the subsequent validation tests employed the BH1 oligonucleotide set.
The performance of the BH1 oligonucleotide-based assay, targeting multicopy sequences in the rRNA, was further compared to a DNA-based assay, which targets a single copy of an essential gene (rnpB) in Listeria species (Mandin et al. 2007; Petrauskene et al. 2017; Yusuf et al. 2010) (Fig. 2). In particular, cell lysates of L. monocytogenes outbreak strain RM2199 were prepared at various tenfold dilutions, ranging from 5000 to 5 cell equivalents (Fig. 2). Aliquots of the cell lysate were subjected to amplification by performing RT-qPCR using the BH1 oligonucleotide-based assay or just PCR using the DNA-based assay targeting rnpB. As shown in Fig. 2, the RNA-based assay with the BH1 probe set resulted in Ct values of 22.7 and 26.9 at the 5000 and 500 cell equivalents, respectively. By contrast, the DNA-based assay resulted in Ct values of 29.1 and 32.1 at the 5000 and 500 cell equivalents, respectively. Interestingly, the BH1 oligonucleotide-based assay still resulted in Ct values of 29.1 and 31.6 at the 50 and 5 cell equivalents, respectively; however, at these lower amounts of Listeria cell equivalents, the DNA-based assay yielded no detectable or Ct values > 35, considered to be a threshold detection limit for gene amplification (McCall et al. 2014). When compared to the DNA-based assay tested, the RNA-based assay, using the BH1 oligonucleotide set, reliably detected cell amounts lower than 5 cells, and these detection sensitivities were statistically significant (Fisher’s Exact Test, P-value = 0.002). These findings revealed that targeting multicopy RNA sequences significantly improved the detection capabilities of the assay for L. monocytogenes, and the results showed that the BH1 oligonucleotide-based assay was found to be at least 100 times more sensitive than the DNA-based assay targeting a single copy gene.
Improved sensitivity of the RNA-based assay when compared to a DNA-based assay. The Y-axis shows the relative fluorescence (ΔRn), the X-axis shows the number of amplification cycles. Yellow symbols represent data obtained with the RNA-based assay, and gray symbols represent data obtained with the DNA-based assay, MicroSEQ™ Listeria monocytogenes Detection kit (Applied Biosystems). Representative data are shown for approximately 5,000 bacterial cells (circles), 500 bacterial cells (triangles), 50 bacterial cells (diamonds), and 5 bacterial cells (squares). The detection threshold is indicated by the dashed line
To further validate the detection capabilities of the assay, the specificity of the BH1 oligonucleotide set was further expanded by testing for Listeria species other than L. monocytogenes. Other additional tested species were L. innocua, L. ivanovii, L. welshimeri, and L. seeligeri, which have been potentially implicated in causing illness to immunocompromised individuals (Korsak and Szuplewska 2016) and have been recovered from ready-to-eat foods or raw/unprocessed foods (Arslan and Özdemir 2020; Guerra et al. 2001; Pesavento et al. 2010; Soriano et al. 2001; Zeinali et al. 2017) as well as in food processing facilities (Huang et al. 2007; Korsak and Szuplewska 2016), and food products (Chen et al. 2009; Pesavento et al. 2010). The inclusivity test was conducted using RNA (100 pg) from Listeria species, corresponding to an estimated 100–150 cell equivalents of RNA per amplification reaction (Glaser et al. 2001; Milner et al. 2001) (Fig. 3). These experiments also examined for cross hybridization of the BH1 oligonucleotide set with excess of non-target RNA (20 ng), which is equivalent to approximately 2 million non-target cells and 4 billion copies of non-target RNA sequences, using an estimated million cell equivalent of RNA per test (Milner et al. 2001) from environmental bacterial species from soil, water, and plant surfaces (Dees et al. 2015; Williams and Marco 2014; Williams et al. 2013) or other foodborne pathogens (Fig. 3). The results indicated a high level of specificity for detecting various Listeria species with Ct values ranging from 19.9 to 25.0, showing a statistically significant difference (Fisher’s Exact Test, P-value = 0.0006) when compared to the lack of detected signals for non-targeted environmental and pathogenic bacterial strains.
Specificity of the oligonucleotide-based assay for target RNA detection. The Y-axis shows the relative fluorescence (ΔRn), the X-axis shows the number of amplification cycles. Representative data are shown for strains of L. grayi (diamonds), L. innocua (circles), L. ivanovii (crosses), L. monocytogenes (squares), L. seeligeri (asterisks), and L. welshimeri (triangles). Data obtained for the negative control and for other gram-positive or gram-negative non-target bacterial strains are presented with lines without symbols. The detection threshold is indicated by the dashed line
Based on recent evidence indicating that the residential bacterial communities are able to persist over time on food production surfaces (Møretrø and Langsrud 2017), additional experiments examined the effect of various amounts of non-target RNA on the efficiency of the amplification of the Listeria sequences. Purified target RNA from L. monocytogenes was co-incubated in the presence of various amounts of excess non-target RNA from B. cereus, a persistent species in food industry environments (Møretrø and Langsrud 2017) and displaying a sequence similarity greater than 90% in the 16S ribosome region (Sallen et al. 1996). As shown in Fig. 4, the results indicated that no significant differences in the Ct values were observed with values ranging from 27.8 to 28.3, corresponding to the signal detected for L. monocytogenes strain RM2199 in the presence of excess non-target RNA from B. cereus strain ATCC 14,579. To determine the effect of the various tested conditions on the amplification efficiency of the target, the slope of the curve at the pre-inflection point was examined (Guescini et al. 2013). Analysis of the amplification curve resulted in no significant change in the slope of the curve under the various conditions tested, and a one-way ANOVA test indicated that the differences between the tested conditions, varying the amounts of non-target template, were not statistically significant (df = 4, F = 3.266, F critical = 5.192). These observations demonstrated that the efficiency of the amplification specific for Listeria was not adversely affected by addition of the non-target template.
Co-incubation of target RNA from L. monocytogenes strain RM2199 in the presence of excess non-target RNA from B. cereus strain ATCC 14,579. The Y-axis shows the relative fluorescence (ΔRn), the X-axis shows the number of amplification cycles. Representative data are shown for 100 fg L. monocytogenes (circles), 100 fg L. monocytogenes plus 100 pg B. cereus (diamonds), 100 fg L. monocytogenes plus 10 pg B. cereus (triangles), 100 fg L. monocytogenes plus 1 pg B. cereus (squares), and asterisks represent data for 100 fg L. monocytogenes plus 100 fg B. cereus (asterisks). The detection threshold is indicated by a dashed line
Design and validation of a continuous flow-through system for Listeria species detection
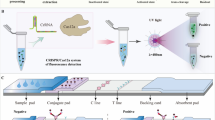
To incorporate the use of the BH1 oligonucleotide-based assay with an improved detection platform, a flow-through system was designed for the real-time surveillance of Listeria species in food processing facilities (Medin et al. 2020). By avoiding the need for sample preparation processes utilizing multistage centrifugation to concentrate the targeted pathogen, the present study replaced the input centrifugation with a flow-through system that included a depth filter to remove large particulate matter while permitting target pathogen to pass through a DNA aptamer-functionalized capture column that specifically bound Listeria cells (Fig. 5). In detail, the filtered sample was introduced into a temperature-controlled chamber (10 °C–12 °C), and fluidic valves moved homogenized sample to a depth filter, which removed large particulate matter present in the collected sample, while still permitting target pathogen to pass through to a microbead capture column that specifically bound Listeria cells (Fig. 5). The specific binding in the capture column was achieved by the use of DNA aptamers (Suh et al. 2014; Suh and Jaykus 2013), which are oligonucleotide molecules that bind to specific epitopes that are presented on the bacterial cell surface and have been proposed for pathogen capture as a low-cost alternative to antibodies (Teng et al. 2016). The use of DNA aptamers enabled Listeria cell capturing without the need for small particle filtering, which can be subject to membrane fouling when isolating bacterial pathogens from food and environmental samples (Ferrari et al. 2019; Kearns et al. 2019; Li et al. 2013; Zhang et al. 2018). To improve the capturing of the Listeria cells with the DNA aptamer (Suh et al. 2014; Suh and Jaykus 2013), additional sequences, a spacer sequence for aptamer extension and a tether sequence for aptamer surface attachment (see Material and Methods)(Quiñones et al. 2020), were designed and added to the aptamer to enable binding to the capture column and prevent the aggregation of the aptamer for efficient release of the target bacterial cell from the aptamer (Medin et al. 2020; Quiñones et al. 2020). A fluidic valve was used to bypass the depth filter, and potential inhibitors were removed as waste. The captured Listeria cells were released from the aptamer-functionalized column, and the liquid sample volume with the Listeria cells was reduced and further subjected to mechanical lysis and amplification with the BH1 oligonucleotide-based assay (Fig. 5). In summary, the designed flow-through system included a sample preparation process with DNA aptamers to specifically capture the targeted Listeria cells away from the contaminating matrix and other non-targeted organisms and inhibitors prior to nucleic acid amplification and analysis.
Schematic diagram of the sample processing steps in the continuous flow-through system. Sample was blended with capture buffer, filtered to remove large particulates, and introduced into a temperature-controlled chamber using a peristaltic pump. Fluidic valves moved homogenized sample to a depth filter, and Listeria cells in the sample were captured with an aptamer-functionalized column. Potential inhibitors were removed as waste, and the cells were subjected to mechanical lysis. The lysed cells were collected and subjected to further amplification by RT-qPCR
As a proof-of-concept, the ability of the flow-through system for efficiently capturing and detecting Listeria cells was further examined. To improve the capture binding efficiency of the Listeria cells, the capture time in the flow-through system was optimized by considering the function of the cross-sectional area of the flow channel and the surface area of the capture surface (Medin et al. 2020). In addition, the aptamer-capture column employed a functionalized surface chemistry based on soda-lime glass surfaces (Medin et al. 2020), one of the least expensive and most-commonly used glass types for many applications. By flowing cell suspensions of L. grayi, a non-pathogenic Listeria species found in raw and ready-to-eat foods (Orsi and Wiedmann 2016; Soriano et al. 2001), through the capture column, the results indicated a 78% capture efficiency of the targeted L. grayi strain RM2208. Given that the tested DNA aptamer may display some affinity for closely related species (Suh et al. 2014; Suh and Jaykus 2013; Teng et al. 2016), Listeria cells captured via aptamer were differentiated using the BH1 oligonucleotide set. To further validate the detection sensitivities of the flow-through system, L. grayi strain RM2208 cell suspensions, consisting of various amounts at or below the infectious dose, based on an estimated infectious dose of Listeria at about 10,000 cells (Pouillot et al. 2016), were determined by traditional plate enumeration on selective solid media. After elution from the capture and ultrasonic mechanical lysis, the results revealed Ct values of 12.3, 16.3, 20.9, and 24.9 for L. grayi cell amounts that were 100 times above the infectious dose, 10 times above the infectious dose, at the infectious dose, and 15 times below the infectious dose, respectively (Fig. 6). Further analysis of the Ct values using the tested serial dilutions of the Listeria cell suspensions were used to calculate the efficiency of the amplification (Kubista et al. 2006), and the analysis revealed that the efficiency of the amplification for detecting Listeria with the flow-through system in conjunction with the BH1 oligonucleotide set had an estimated amplification efficiency of 0.9235 (R = 0.99955; slope -3.52). These findings indicated that this integrated flow-through system has the capabilities to accurately and sensitively detect Listeria species.
Detection sensitivity of the oligonucleotide-based assay with the continuous flow-through system. The Y-axis shows the relative fluorescence (ΔRn), the X-axis shows the number of amplification cycles. Representative data are shown for samples containing L. grayi strain RM2208 at equivalent amounts of 100 times above the Listeria infectious dose (circles), at 10 times above the Listeria infectious dose (diamonds), at the Listeria infectious dose (triangles), at 15 times below the Listeria infectious dose (squares). The negative control is indicated by the gray line. The detection threshold is indicated by the dashed line
Conclusions
In conclusion, the present study developed a singleplex RT-qPCR assay using the BH1 oligonucleotide set for the accurate detection of multiple Listeria species. Previously published assays for Listeria species detection employed qPCR and were developed by targeting iap, plcB, and scrA genes, required for virulence and regulatory functions, as well as putative internalin and oxidoreductase, and N-acetylmuramidase proteins (Chen et al. 2017; Hage et al. 2014; Hein et al. 2001; Hitchins et al. 2017). Other qPCR assays have been documented, which target the 23S ribosomal DNA or non-coding RNA specific to the Listeria genus (Chen et al. 2017; Petrauskene et al. 2017; Rodríguez-Lázaro et al. 2004). The disadvantage of these published assays is the requirement of multiple set of primers and probes to be multiplexed per amplification reaction to enable the detection of various Listeria species.
As an alternative to DNA-based methods by employing qPCR, the present study developed a RT-qPCR assay that detected rRNA in Listeria species. The advantage of using rRNA as the target in Listeria species identification is that the rRNA genes are present in this pathogen at all stages of growth, accumulate mutations at a slow constant rate, and contain sequences that enable discrimination of this targeted pathogen at the genus level (Espejo and Plaza 2018; Milner et al. 2001). Additionally, analysis of the Listeria genome revealed large amounts of rRNA (600–25,000 copies per cell) (Milner et al. 2001), which makes rRNA a better target for a sensitive detection at low cell concentrations when compared with one copy of genomic DNA for a targeted gene. The explosive use of genome sequencing has resulted in an increase in the growth of data (Land et al. 2015; Sayers et al. 2020), which has consequently improved the in silico sequence analyses for the identification of rRNA sequences that are highly specific for the targeted pathogen. In the present study, the designed singleplex RT-qPCR assay using the BH1 oligonucleotide assay, targeting 23S rRNA, resulted in the reliable detection of less than 5 Listeria cells, and this detection was at least 100 times more sensitive than the DNA-based assay targeting a single copy gene. Furthermore, the BH1 oligonucleotide assay results showed no cross reactivity for detecting non-Listeria strains in the exclusivity test even at template amounts that were 100-fold when compared to the targeted Listeria species.
The use of enrichment in routine surveillance testing is traditionally performed to amplify the target organism exponentially by as much as a millionfold so that detection is possible (Law et al. 2015). To improve the detection of pathogens from samples, various reports have documented on using microfiltration to reduce large samples to a small volume (Li et al. 2013). However, small particle filtering can be subject to membrane fouling when isolating bacterial pathogens from food and environmental samples, resulting in reduced levels of detection sensitivities (Kearns et al. 2019; Li et al. 2013; Zhang et al. 2018). In the present study, the flow-through system did not use membrane filtration but instead used an adaptation of depth filtering, a processing stage previously used for preventing filter clogging (Murakami 2012), followed by the use of an aptamer-functionalized column for successfully capturing and concentrating the targeted Listeria cells from samples. The findings from the present study indicated the flow-through system accurately detected Listeria cells at cell concentrations that were 15 times below the infectious dose. Due to the non-uniform distribution of microorganisms, current methods employing small sample sizes can result in undetected pathogens, and the lack of accurate detection of the tested foodborne pathogen can lead to subsequent outbreaks (Capobianco et al. 2021; Kearns et al. 2019). To continue to meet the needs of the food safety industry and regulators (Kaplan et al. 2014; Kuiper and Paoletti 2015), further research is aimed at optimizing the high binding efficiency of the flow-through system, documented in the present study, to optimize the rapid, low-cost capture of Listeria species using increased sample size over a range of environmental and food samples. Moreover, recent preliminary evidence demonstrated that the flow-through system accurately detected Listeria at low cell concentrations, ranging from 3.5 CFU/ml to 1000 CFU/ml, in samples collected at a food processing facility (Quiñones et al. 2019, 2020) and these challenging samples were characterized by containing contaminants commonly found in soils and agricultural products, including debris and humic acids, which are known inhibitors of nucleic acid amplification (Law et al. 2015; Opel et al. 2010; Schrader et al. 2012). These observations indicate that the high capture efficiency and strong binding affinity of aptamers towards the targeted Listeria cells enable aggressive washes to remove inhibitors and substances that negatively impact nucleic acid amplification and would thus enable the detection of Listeria cells in other types of samples such as complex food matrices, known to contain PCR inhibitory substances (Schrader et al. 2012). Future studies will also explore the increased sampling size of the method, described in the present study, to improve the statistical significance of the testing procedure and to enable a sampling process that is more accurate and representative of the entire agricultural field, providing an added value to the food industry.
Availability of data and material
The material analyzed during the current study is available from the corresponding author on reasonable request.
References
Arslan S, Özdemir F (2020) Prevalence and antimicrobial resistance of Listeria species and molecular characterization of Listeria monocytogenes isolated from retail ready-to-eat foods. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnaa006
Bettelheim KA, Evangelidis H, Pearce JL, Sowers E, Strockbine NA (1993) Isolation of a Citrobacter freundii strain which carries the Escherichia coli O157 antigen. J Clin Microbiol 31:760–761. https://doi.org/10.1128/jcm.31.3.760-761.1993
Brandl MT, Mandrell RE (2002) Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. J Appl Environ Microbiol 68:3614–3621. https://doi.org/10.1128/AEM.68.7.3614-3621.2002
Brouillette R, Aggen D, Borchert B, Buckman K, Domanico M, Fraser-Heaps J, Freier T, Hayman M, Jackson T, Kataoka A, Meyer J, Shoaf E, Stone W (2014) Listeria monocytogenes guidance on environmental monitoring and corrective actions in at-risk foods. Grocery Manufacturers Association, Washington
Buchanan RL, Gorris LGM, Hayman MM, Jackson TC, Whiting RC (2017) A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75:1–13. https://doi.org/10.1016/j.foodcont.2016.12.016
Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P, Frangeul L, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakaborty T, Charbit A, Chétouani F, Couvé E, De Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durand L, Dusurget O, Entian KD, Fsihi H, Garcia-Del Portillo P, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kärst U, Kreft J, Kuhn M, Kurapkat G, Madueño E, Maitournam A, Mata Vicente J, Ng E, Nordsiek G, De Pablos B, Pérez-Diaz JC, Remmel B, Rose M, Schlueter T, Simoes N, Vázquez-Boland JA, Voss H, Wehland J (2003) Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: Clues for evolution and pathogenicity. FEMS Immunol Med Microbiol 35:207–213. https://doi.org/10.1016/S0928-8244(02)00448-0
Capobianco JA, Armstrong CM, Lee J, Gehring AG (2021) Detection of pathogenic bacteria in large volume food samples using an enzyme-linked immunoelectrochemical biosensor. Food Control. https://doi.org/10.1016/j.foodcont.2020.107456
Charlermroj R, Makornwattana M, Phuengwas S, Meerak J, Pichpol D, Karoonuthaisiri N (2019) DNA-based bead array technology for simultaneous identification of eleven foodborne pathogens in chicken meat. Food Control 101:81–88. https://doi.org/10.1016/j.foodcont.2019.02.014
Chen J, Zhang X, Mei L, Jiang L, Fang W (2009) Prevalence of Listeria in Chinese food products from 13 provinces between 2000 and 2007 and virulence characterization of Listeria monocytogenes isolates. Foodborne Pathog Dis 6:7–14. https://doi.org/10.1089/fpd.2008.0139
Chen JQ, Healey S, Regan P, Laksanalamai P, Hu Z (2017) PCR-based methodologies for detection and characterization of Listeria monocytogenes and Listeria ivanovii in foods and environmental sources. Food Sci Hum Well 6:39–59. https://doi.org/10.1016/j.fshw.2017.03.001
Churchill KJ, Sargeant JM, Farber JM, O’connor AM, (2019) Prevalence of Listeria monocytogenes in select ready-to-eat foods—deli meat, soft cheese, and packaged salad: A systematic review and meta-analysis. J Food Prot 82:344–357. https://doi.org/10.4315/0362-028X.JFP-18-158
Cooley MB, Miller WG, Mandrell RE (2003) Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. J Appl Environ Microbiol 69:4915–4926. https://doi.org/10.1128/AEM.69.8.4915-4926.2003
Cooley M, Carychao D, Crawford-Miksza L, Jay MT, Myers C, Rose C, Keys C, Farrar J, Mandrell RE (2007) Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS ONE. https://doi.org/10.1371/journal.pone.0001159
Cooley MB, Jay-Russell M, Atwill ER, Carychao D, Nguyen K, Quiñones B, Patel R, Walker S, Swimley M, Pierre-Jerome E, Gordus AG, Mandrell RE (2013) Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS ONE. https://doi.org/10.1371/journal.pone.0065716
Cooley MB, Quiñones B, Oryang D, Mandrell RE, Gorski L (2014) Prevalence of Shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2014.00030
Dees MW, Lysøe E, Nordskog B, Brurberg MB (2015) Bacterial communities associated with surfaces of leafy greens: Shift in composition and decrease in richness over time. J Appl Environ Microbiol 81:1530–1539. https://doi.org/10.1128/AEM.03470-14
Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P (2004) Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. https://doi.org/10.1128/JCM.42.8.3819-3822.2004
Espejo RT, Plaza N (2018) Multiple ribosomal RNA operons in bacteria; Their concerted evolution and potential consequences on the rate of evolution of their 16S rRNA. Front Microbiol. https://doi.org/10.3389/fmicb.2018.01232
Estrada EM, Hamilton AM, Sullivan GB, Wiedmann M, Critzer FJ, Strawn LK (2020) Prevalence, persistence, and diversity of Listeria monocytogenes and Listeria species in produce packinghouses in three U.S. States J Food Prot 83:277–286. https://doi.org/10.4315/0362-028X.JFP-19-411
Fagerquist CK, Zaragoza WJ (2018) Proteolytic surface-shaving and serotype-dependent expression of SPI-1 invasion proteins in Salmonella enterica subspecies enterica. Front Nutr. https://doi.org/10.3389/fnut.2018.00124
Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE (2005) Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci U S A 102:11064–11069. https://doi.org/10.1073/pnas.0504930102
Ferrari S, Frosth S, Svensson L, Fernström LL, Skarin H, Hansson I (2019) Detection of Campylobacter spp. in water by dead-end ultrafiltration and application at farm level. J Appl Microbiol 127:1270–1279. https://doi.org/10.1111/jam.14379
Fisher RA (1935) The Logic of Inductive Inference. J R Stat Soc 98:39–82. https://doi.org/10.2307/2342435
Friedman M, Henika PR, Levin CE, Mandrell RE, Kozukue N (2006) Antimicrobial activities of tea catechins and theaflavins and tea extracts against Bacillus cereus. J Food Prot 69:354–361. https://doi.org/10.4315/0362-028X-69.2.354
Gasanov U, Hughes D, Hansbro PM (2005) Methods for the isolation and identification of Listeria spp. and Listeria monocytogenes: A review. FEMS Microbiol Rev 29:851–875. https://doi.org/10.1016/j.femsre.2004.12.002
Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, De Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Garcia-Del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueño E, Maitournam A, Mata Vicente J, Ng E, Nedjari H, Nordsiek G, Novella S, De Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vázquez-Boland JA, Voss H, Wehland J, Cossart P (2001) Comparative genomics of Listeria species. Science 294:849–852. https://doi.org/10.1126/science.1063447
Gorski L, Flaherty D, Mandrell RE (2006) Competitive fitness of Listeria monocytogenes serotype 1/2a and 4b strains in mixed cultures with and without food in the U.S. Food and Drug Administration enrichment protocol. J Appl Environ Microbiol 72:776–783. https://doi.org/10.1128/AEM.72.1.776-783.2006
Guerra MM, McLauchlin J, Bernardo FA (2001) Listeria in ready-to-eat and unprocessed foods produced in Portugal. Food Microbiol 18:423–429. https://doi.org/10.1006/fmic.2001.0421
Guescini M, Sisti D, Rocchi MBL, Panebianco R, Tibollo P, Stocchi V (2013) Accurate and precise DNA quantification in the presence of different amplification efficiencies using an improved Cy0 method. PLoS ONE. https://doi.org/10.1371/journal.pone.0068481
Hage E, Mpamugo O, Ohai C, Sapkota S, Swift C, Wooldridge D, Amar CFL (2014) Identification of six Listeria species by real-time PCR assay. Lett Appl Microbiol 58:535–540. https://doi.org/10.1111/lam.12223
Hein I, Klein D, Lehner A, Bubert A, Brandl E, Wagner M (2001) Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res Microbiol 152:37–46. https://doi.org/10.1016/S0923-2508(00)01166-9
Hitchins AD, Jinneman K, Chen Y (2017) BAM Chapter 10: Detection of Listeria monocytogenes in foods and environmental samples, and enumeration of Listeria monocytogenes in foods. https://www.fda.gov/food/laboratory-methods-food/bam-detection-and-enumeration-listeria-monocytogenes. Accessed 03/04/2020
Hoffmann S, Scallan Walter E (2019) Acute complications and sequelae from foodborne infections: Informing priorities for cost of foodborne illness estimates. Foodborne Pathog Dis. https://doi.org/10.1089/fpd.2019.2664
Hoffmann S, Batz MB, Morris JG Jr (2012) Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J Food Prot 75:1292–1302. https://doi.org/10.4315/0362-028X.JFP-11-417
Hoffmann S, Maculloch B, Batz M (2015) Economic burden of major foodborne illnesses acquired in the United States. EIB-140, U.S. Department of Agriculture, Economic Research Service, May 2015. https://www.ers.usda.gov/webdocs/publications/43984/52807_eib140.pdf
Huang B, Eglezos S, Heron BA, Smith H, Graham T, Bates J, Savill J (2007) Comparison of multiplex PCR with conventional biochemical methods for the identification of Listeria spp. isolates from food and clinical samples in Queensland. Australia J Food Prot 70:1874–1880. https://doi.org/10.4315/0362-028X-70.8.1874
Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D’Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91. https://doi.org/10.1038/nature01582
Kaplan RM, Chambers DA, Glasgow RE (2014) Big data and large sample size: A cautionary note on the potential for bias. J Clin Transl Sci 7:342–346. https://doi.org/10.1111/cts.12178
Kearns EA, Gustafson RE, Castillo SM, Alnughaymishi H, Lim DV, Ryser ET (2019) Rapid large-volume concentration for increased detection of Escherichia coli O157:H7 and Listeria monocytogenes in lettuce wash water generated at commercial facilities. Food Control 98:481–488. https://doi.org/10.1016/j.foodcont.2018.12.006
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Korsak D, Szuplewska M (2016) Characterization of nonpathogenic Listeria species isolated from food and food processing environment. Int J Food Microbiol 238:274–280. https://doi.org/10.1016/j.ijfoodmicro.2016.08.032
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N (2006) The real-time polymerase chain reaction. Mol Aspects Med 27:95–125. https://doi.org/10.1016/j.mam.2005.12.007
Kuiper HA, Paoletti C (2015) Food and feed safety assessment: The importance of proper sampling. J AOAC Int 98:252–258. https://doi.org/10.5740/jaoacint.15-007
Kutyavin IV (2008) Use of base-modified duplex-stabilizing deoxynucleoside 5′-triphosphates to enhance the hybridization properties of primers and probes in detection polymerase chain reaction. Biochemistry 47:13666–13673. https://doi.org/10.1021/bi8017784
Kyle JL, Cummings CA, Parker CT, Quiñones B, Vatta P, Newton E, Huynh S, Swimley M, Degoricija L, Barker M, Fontanoz S, Nguyen K, Patel R, Fang R, Tebbs R, Petrauskene O, Furtado M, Mandrell RE (2012) Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J Bacteriol 194:1885–1896. https://doi.org/10.1128/JB.00120-12
Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW (2015) Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–161. https://doi.org/10.1007/s10142-015-0433-4
Law JW, Ab Mutalib NS, Chan KG, Lee LH (2015) Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front Microbiol. https://doi.org/10.3389/fmicb.2014.00770
Li X, Ximenes E, Amalaradjou MAR, Vibbert HB, Foster K, Jones J, Liu X, Bhunia AK, Ladisch MR (2013) Rapid sample processing for detection of food-borne pathogens via cross-flow microfiltration. J Appl Environ Microbiol 79:7048–7054. https://doi.org/10.1128/AEM.02587-13
Liu Y, Xu A, Fratamico PM, Sommers CH, Rotundo L, Boccia F, Jiang Y, Ward TJ (2018) Draft whole-genome sequences of seven Listeria monocytogenes strains with variations in virulence and stress responses. Microbiol Resour Announc. https://doi.org/10.1128/MRA.01038-18
Livezey K, Kaplan S, Wisniewski M, Becker MM (2013) A new generation of food-borne pathogen detection based on ribosomal RNA. Annu Rev Food Sci Technol 4:313–325. https://doi.org/10.1146/annurev-food-050412-104448
Longhi C, Ammendolia MG, Conte MP, Seganti L, Iosi F, Superti F (2014) Listeria ivanovii ATCC 19119 strain behaviour is modulated by iron and acid stress. Food Microbiol 42:66–71. https://doi.org/10.1016/j.fm.2014.02.015
Mandin P, Geissmann T, Cossart P, Repoila F, Vergassola M (2007) Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 35:962–974. https://doi.org/10.1093/nar/gkl1096
McCall MN, McMurray HR, Land H, Almudevar A (2014) On non-detects in qPCR data. Bioinformatics 30:2310–2316. https://doi.org/10.1093/bioinformatics/btu239
Medin DL, De Guzman VS, Panchal Z (2020) System and method for detecting and monitoring pathogens. USA Patent United States Patent and Trademark Office, Patent Application Number 2020/043112
Mehta CR, Patel NR (1986) Algorithm 643. FEXACT: a FORTRAN subroutine for Fisher’s exact test on unordered r×c contingency tables. ACM Trans Math Softw. 12:154–161
Milner MG, Saunders JR, McCarthy AJ (2001) Relationship between nucleic acid ratios and growth in Listeria monocytogenes. Microbiology 147:2689–2696. https://doi.org/10.1099/00221287-147-10-2689
Møretrø T, Langsrud S (2017) Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr Rev Food Sci Food Saf 16:1022–1041. https://doi.org/10.1111/1541-4337.12283
Murakami T (2012) Filter-based pathogen enrichment technology for detection of multiple viable foodborne pathogens in 1 day. J Food Prot 75:1603–1610. https://doi.org/10.4315/0362-028X.JFP-12-039
Nielsen HB, Wernersson R, Knudsen S (2003) Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res 31:3491–3496. https://doi.org/10.1093/nar/gkg622
Niemira BA, Fan X, Sokorai KJB, Sommers CH (2003) Ionizing radiation sensitivity of Listeria monocytogenes ATCC 49594 and Listeria innocua ATCC 51742 inoculated on endive (Cichorium endiva). J Food Prot 66:993–998. https://doi.org/10.4315/0362-028X-66.6.993
Opel KL, Chung D, McCord BR (2010) A study of PCR inhibition mechanisms using real time PCR. J Forensic Sci 55:25–33. https://doi.org/10.1111/j.1556-4029.2009.01245.x
Orsi RH, Wiedmann M (2016) Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl Microbiol Biotechnol 100:5273–5287. https://doi.org/10.1007/s00253-016-7552-2
Pesavento G, Ducci B, Nieri D, Comodo N, Lo Nostro A (2010) Prevalence and antibiotic susceptibility of Listeria spp. isolated from raw meat and retail foods. Food Control 21:708–713. https://doi.org/10.1016/j.foodcont.2009.10.012
Petrauskene O, Cummings C, Vatta P, Tebbs R, Balachandran P, Zoder P, Wong L (2017) Detection of Listeria species in food and environmental samples, methods and compositions thereof. USA Patent United States Patent and Trademark Office, Patent Number 9,546,405 B2
Pouillot R, Klontz KC, Chen Y, Burall LS, Macarisin D, Doyle M, Bally KM, Strain E, Datta AR, Hammack TS, Van Doren JM (2016) Infectious dose of Listeria monocytogenes in outbreak linked to ice cream, United States, 2015. Emerg Infect Dis 22:2113–2119. https://doi.org/10.3201/eid2212.160165
Quiñones B, Swimley MS, Taylor AW, Dawson ED (2011) Identification of Escherichia coli O157 by using a novel colorimetric detection method with DNA microarrays. Food Path Dis 8:705–711. https://doi.org/10.1089/fpd.2010.0753
Quiñones B, Swimley MS, Narm KE, Patel RN, Cooley MB, Mandrell RE (2012) O-antigen and virulence profiling of Shiga toxin-producing Escherichia coli by a rapid and cost-effective DNA microarray colorimetric method. Front Cell Infect Microbiol 2:61. https://doi.org/10.3389/fcimb.2012.00061
Quiñones B, Lee BG, Martinsky TJ, Yambao JC, Haje PK, Schena M (2017) Sensitive genotyping of foodborne-associated human noroviruses and hepatitis A virus using an array-based platform. Sensors 17:1–16. https://doi.org/10.3390/s17092157
Quiñones B, De Guzman V, Yambao JC, Medin DL, Lee BG (2019) Development of an integrated detection platform for the in-process surveillance of Listeria spp. in environmental monitoring samples. Paper presented at the International Association for Food Protection Annual Meeting, Louisville, KY
Quiñones B, Lee BG, Yambao JC (2020) Listeria detection. United States Patent and Trademark Office, Patent Application Number 16/815,323
Rocourt J, Boerlin P, Grimont F, Jacquet C, Piffaretti JC (1992) Assignment of Listeria grayi and Listeria murrayi to a single species, Listeria grayi, with a revised description of Listeria grayi. Int J Syst Evol Microbiol 42:171–174. https://doi.org/10.1099/00207713-42-1-171
Rodríguez-Lázaro D, Hernández M, Pla M (2004) Simultaneous quantitative detection of Listeria spp. and Listeria monocytogenes using a duplex real-time PCR-based assay. FEMS Microbiol Lett 233:257–267. https://doi.org/10.1016/j.femsle.2004.02.018
Rosimin AA, Kim MJ, Joo IS, Suh SH, Kim KS (2016) Simultaneous detection of pathogenic Listeria including atypical Listeria innocua in vegetables by a quadruplex PCR method. LWT - Food Sci Technol 69:601–607. https://doi.org/10.1016/j.lwt.2016.02.007
Ryu J, Park SH, Yeom YS, Shrivastav A, Lee SH, Kim YR, Kim HY (2013) Simultaneous detection of Listeria species isolated from meat processed foods using multiplex PCR. Food Control 32:659–664. https://doi.org/10.1016/j.foodcont.2013.01.048
Sallen B, Rajoharison A, Desvarenne S, Quinn F, Mabilat C (1996) Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int J Syst Bacteriol 46:669–674. https://doi.org/10.1099/00207713-46-3-669
Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I (2020) GenBank Nuc Acids Res 48:D84–D86. https://doi.org/10.1093/nar/gkz956
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM (2011) Foodborne illness acquired in the United States-Major pathogens. Emerg Infect Dis 17:7–15. https://doi.org/10.3201/eid1701.P11101
Schrader C, Schielke A, Ellerbroek L, Johne R (2012) PCR inhibitors - occurrence, properties and removal. J Appl Microbiol 113:1014–1026. https://doi.org/10.1111/j.1365-2672.2012.05384.x
Shanks RMQ, Dashiff A, Alster JS, Kadouri DE (2012) Isolation and identification of a bacteriocin with antibacterial and antibiofilm activity from Citrobacter freundii. Arch Microbiol 194:575–587. https://doi.org/10.1007/s00203-012-0793-2
Soriano JM, Rico H, Moltó JC, Mañes J (2001) Listeria species in raw and ready-to-eat foods from restaurants. J Food Prot 64:551–553. https://doi.org/10.4315/0362-028X-64.4.551
Sowers BA, Songster MF, Smith KE, Snyder DL, Cook RM (2005) RealTimeDesign™ Software - An Advanced Web-based Program for Real-time PCR Sequence Design. In: 2nd International qPCR Event, Freising-Weihenstephan, Germany
Sowers BA, Songster MF, Smith KE, Snyder DL, Cook RM (2006) RealTimeDesign™ - Free Web-based Software For Dual-labeled and Amplifluor® Design In: 20th Anniversary San Diego Conference DNA Probe Technology, San Diego
Suh SH, Jaykus LA (2013) Nucleic acid aptamers for capture and detection of Listeria spp. J Biotechnol 167:454–461. https://doi.org/10.1016/j.jbiotec.2013.07.027
Suh SH, Dwivedi HP, Choi SJ, Jaykus LA (2014) Selection and characterization of DNA aptamers specific for Listeria species. Anal Biochem 459:39–45. https://doi.org/10.1016/j.ab.2014.05.006
Teng J, Yuan F, Ye Y, Zheng L, Yao L, Xue F, Chen W, Li B (2016) Aptamer-based technologies in foodborne pathogen detection. Front Microbiol. https://doi.org/10.3389/fmicb.2016.01426
The United Fresh Food Safety & Technology Council (2018) Guidance on enviromnental monitoring and control of Listeria for the fresh produce industry, 2nd edn. United Fresh Produce Association, Washington
Waleh NS, Ingraham JL (1976) Pyrimidine ribonucleoside monophosphokinase and the mode of RNA turnover in Bacillus subtilis. Arch Microbiol 110:49–54. https://doi.org/10.1007/BF00416968
Wei S, Daliri EBM, Chelliah R, Park BJ, Lim JS, Baek MA, Nam YS, Seo KH, Jin YG, Oh DH (2019) Development of a multiplex real-time PCR for simultaneous detection of Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus in food samples. J Food Saf. https://doi.org/10.1111/jfs.12558
Williams TR, Marco ML (2014) Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. mBio. https://doi.org/10.1128/mBio.01564-14
Williams TR, Moyne AL, Harris LJ, Marco ML (2013) Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS ONE. https://doi.org/10.1371/journal.pone.0068642
Woese CR (1987) Bacterial evolution. Microbiol Rev 51:221–271. https://doi.org/10.1128/mmbr.51.2.221-271.1987
Yusuf D, Marz M, Stadler PF, Hofacker IL (2010) Bcheck: A wrapper tool for detecting RNase P RNA genes. BMC Genom. https://doi.org/10.1186/1471-2164-11-432
Zeinali T, Jamshidi A, Bassami M, Rad M (2017) Isolation and identification of Listeria spp. in chicken carcasses marketed in northeast of Iran. Int Food Res J 24:881–887
Zhang Y, Xu CQ, Guo T, Hong L (2018) An automated bacterial concentration and recovery system for pre-enrichment required in rapid Escherichia coli detection. Sci Rep. https://doi.org/10.1038/s41598-018-35970-8
Zoellner C, Ceres K, Ghezzi-Kopel K, Wiedmann M, Ivanek R (2018) Design elements of Listeria environmental monitoring programs in food processing facilities: A scoping review of research and guidance materials. Compr Rev Food Sci Food Saf 17:1156–1171. https://doi.org/10.1111/1541-4337.12366
Acknowledgements
This material was based in part upon work supported by the United States Department of Agriculture (USDA), Agricultural Research Service, CRIS project 2030-42000-051-00D under the Cooperative Research and Development Agreement No. 58-2030-5-008. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Funding
This work was supported by the United States Department of Agriculture, Agricultural Research Service, CRIS Project Number 2030-42000-051-00D.
Author information
Authors and Affiliations
Contributions
BQ, VSDG and DLM conceived and designed the experiments. BQ, VSDG and DLM provided important advice, reagents, and analysis tools to the design of the test. BGL and DLM conducted bioinformatics analyses. JCY, VSDG and BGL performed the experiments. BQ, JCY, VSDG, BGL and DLM analyzed the data. BQ, JCY and BGL wrote the manuscript. All authors read, reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
Authors BQ, JCY and BGL have no relevant financial or non-financial interests to declare that are relevant to the content of this article. Authors VSDG and DLM are employed by Snap DNA-BioNEMS, Inc. (Mountain View, CA, USA).
Consent for publication
All authors read and approved the final version of the manuscript for publication.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Quiñones, B., Yambao, J.C., De Guzman, V.S. et al. Genomic analysis of high copy-number sequences for the targeted detection of Listeria species using a flow-through surveillance system. Arch Microbiol 203, 3667–3682 (2021). https://doi.org/10.1007/s00203-021-02388-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02388-2