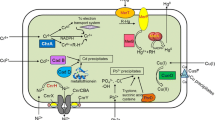
Abstract
Mercury pollution has emerged as a major problem in industrialized zones and presents a serious threat to environment and health of local communities. Effectiveness and wide distribution of mer operon by horizontal and vertical gene transfer in its various forms among large community of microbe reflect importance and compatibility of this mechanism in nature. This review specifically describes mer operon and its generic molecular mechanism with reference to the central role played by merA gene and its related gene products. The combinatorial action of merA and merB together maintains broad spectrum mercury detoxification system for substantial detoxification of mercurial compounds. Feasibility of mer operon to coexist with antibiotic resistance gene (amp r, kan r, tet r) clusters enables extensive adaptation of bacterial species to adverse environment. Flexibility of the mer genes to exist as intricate part of chromosome, plasmids, transposons, and integrons enables high distribution of these genes in wider microbial gene pool. Unique ability of this system to manipulate oligodynamic property of mercurial compounds for volatilization of mercuric ions (Hg2+) makes it possible for a wide range of microbes to tolerate mercury-mediated toxicity.

Similar content being viewed by others
Abbreviations
- DGM:
-
Dissolved gaseous mercury
- HGT:
-
Horizontal gene transfer
- MeHg:
-
Methylmercury
- O/P:
-
Operator/promoter
- PCR:
-
Polymerase chain reaction
- PHB:
-
Polyhydroxybutyrate
References
Abou-Shanab RA, van Berkum P, Angle JS (2007) Heavy metal resistance and genotypic analysis of metal resistances genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 68:360–367
Baath E (1989) Effects of heavy metals in soil on microbial processes and populations. Water Air Soil Pollut 47:335–379
Barkay T (1987) Adaptation of aquatic microbial communities to Hg+2 stress. Appl Environ Microbiol 53:2725–2732
Barkay T, Miller SM, Summers AO (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27:355–384
Barkay T, Kritee K, Boyd E, Geesey G (2010) A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ Microbiol 12:2904–2917
Barnes HL, Seward TM (1997) Geothermal systems and mercury deposits. In: Barnes HL (ed) Geochemistry of hydrothermal ore deposits, 3rd edn. Wiley, NewYork, pp 699–736
Bogdanova E, Mindlin S, Pakrava E, Kocur M, Rouch D (1992) Mercuric reductase in enviromental gram-positive bacteria sensitive to mercury. FEMS Microbiol Lett 97:95–100
Boni MF, Feldman MW (2005) Evolution of antibiotic resistance by human and bacterial niche construction. Evolution 59:477–491
Chatziefthimiou AD, Crespo-Medina M, Wang Y et al (2007) The isolation and initial characterization of mercury resistant chemolithotrophic thermophilic bacteria from mercury rich geothermal springs. Extremophiles 11:469–479
Chien MF, Narita M, Lin KH, Matsui K, Huang CC, Endo G (2010) Organomercurials removal by heterogeneous merB genes harboring bacterial strains. J Biosci Bioeng 110:94–98
de Lipthay JR, Rasmussen LD, Oregaard G, Simonsen K, Bahl MI, Kroer N, Sørensen SJ (2008) Acclimation of subsurface microbial communities to mercury. FEMS Microbiol Ecol 65:145–155
De J, Ramaiah N, Vardanyan L (2008) Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar Biotechnol 10:471–477
Fantozzi L, Ferrara R, Frontini FP, Dini F (2009) Dissolved gaseous mercury production in the dark: evidence for the fundamental role of bacteria in different types of Mediterranean water bodies. Sci Total Environ 407:917–924
Fenchel T, King GH, Blackburn TH (1999) Bacterial biogeochemistry. The ecophysiology of mineral cycling. Int Microbiol 2:201–204
Foster TJ (1983) Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev 47:361–409
Grier N (1977) Mercurials-inorganic and organic in disinfection, sterilization, and preservation, 2nd edn. Lea and Febiger, Philadelphia, pp 361–385
Griffin HG, Foster TJ, Silver S, Misra TK (1987) Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinant of plasmid pDU1358. Proc Natl Acad Sci USA 84:3112–3116
Hattemer AJ (1954) Oligodynamic effects of heavy metals. Zahnarztl Rundsch 63:431–436
Huang CC, Chen MW, Hsieh JL, Lin WH, Chen PC, Chien LF (2006) Expression of mercuric reductase from Bacillus megaterium MB1 in eukaryotic microalga Chlorella sp. DT: an approach for mercury phytoremediation. Appl Microbiol Biotechnol 72:197–205
Huang X, Sillanpää M, Duo B, Gjessing ET (2008) Water quality in the Tibetan plateau: metal contents of four selected rivers. Environ Pollut 156:270–277
Huang X, Sillanpää M, Gjessing E, Vogt RD (2009) Water quality in the Tibetan plateau: major ions and trace elements in the headwaters of four major Asian rivers. Sci Tot Environ 407:6242–6254
Huang X, Sillanpää M, Gjessing ET, Peräniemi S, Vogt RD (2011) Water quality in the southern Tibetan plateau: chemical evaluation of the river Yarlung Tsangpo (Brahmaputra). River Res Appl 27:113–121
Inoue C, Kusano T, Silver S (1996) Mercuric ion uptake by Escherichia coli cells producing Thiobacillus ferrooxidans MerC. Biosci Biotechnol Biochem 60:1289–1292
Janssen PJ, van Houdt R, Moors H, Monsieurs P, Morin N et al (2010) The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 5:e10433. doi:10.1371/journal.pone.0010433
Kim EH, Mason RP, Porter ET, Soulen HJ (2006) The impact of resuspension on sediment mercury dynamics, and methylmercury production and fate: a mesocosm study. Mar Chem 102:300–315
Lal D, Lal R (2010) Evolution of mercuric reductase (merA) gene: a case of horizontal gene transfer. Mikrobiologiia 79:524–531
Liebert CA, Wireman J, Smith T, Summers AO (1997) Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl Environ Microbiol 63:1066–1076
Lyyra S, Meagher RB, Kim T et al (2007) Coupling two mercury resistance genes in eastern cottonwood enhances the processing of organomercury. Plant Biotechnol J 5:254–262
Martins AS, Silva de Jesus M, Lacerda M, Moreira JC, Filgueiras ALL, Barrocas PRG (2008) A conservative region of the mercuric reductase gene (merA) as a molecular marker of bacterial mercury resistance. Braz J Microbiol 39:307–310
Moore B (1960) A new screen test and selective medium for the rapid detection of epidemic strains of Staphylococcus aureus. Lancet 2:453–458
Müller AK, Westergaard K, Christensen S, Sorensen SJ (2001) The effect of long-term mercury pollution on the soil microbial community. FEMS Microbiol Ecol 36:11–19
Murtaza I, Dutt A, Mushtaq D, Ali A (2005) Molecular cloning and genetic analysis of functional merB gene from Indian isolates of Escherichia coli. Curr Microbiol 51:297–302
Nagata T, Kiyono M, Pan-Hou H (2006) Accumulation of mercury in transgenic tobacco expressing bacterial polyphosphate. Biol Pharm Bull 29:2350–2353
Nascimento AMA, Chartone-Souza E (2003) Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet Mol Res 2:92–101
National Research Council (2000) Toxicological effects of methylmercury. National Academy Press, Washington, pp 147–246
Osborn AM, Bruce KD, Strike P, Ritchie DA (1997) Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev 4:239–262
Ramond JB, Berthe T, Duran R, Petit F (2009) Comparative effects of mercury contamination and wastewater effluent input on gram-negative merA gene abundance in mudflats of an anthropized estuary (Seine, France): a microcosm approach. Res Microbiol 160:10–18
Rasmussen LD, Sørensen SJ (2001) Effects of mercury contamination on the culturable heterotrophic, functional and genetic diversity of the bacterial community in soil. FEMS Microbiol Ecol 36:1–9
Rassaei L, Sillanpää M, Edler KJ, Marken F (2009) Electrochemically active mercury nanodroplets trapped in a carbon nanoparticle—chitosan matrix. Electroanalysis 21:261–266
Roberts MC, Leroux BG, Sampson J, Luis HS, Bernardo M, Leitão J (2008) Dental amalgam and antibiotic- and/or mercury-resistant bacteria. J Dent Res 87:475–479
Rojas LA, Yanez C, Gonzalez M, Lobos S, Smalla K, Seeger M (2011) Characterization of the metabolically modified heavy metal-resistant Cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS ONE 6(3):e17555. doi:10.1371/journal.pone.0017555
Rudrick JT, Bawdon RE, Guss SP (1985) Determination of mercury and organomercurial resistance in obligate anaerobic bacteria. Can J Microbiol 31:276–281
Ruiz ON, Daniell H (2009) Genetic engineering to enhance mercury phytoremediation. Curr Opin Biotechnol 20:213–219
Ruta L, Paraschivescu C, Matache M et al (2010) Removing heavy metals from synthetic effluents using ‘‘kamikaze’’ Saccharomyces cerevisiae cells. Appl Microbiol Biotechnol 85:763–771
Sadhukhan PC, Ghosh S, Chaudhuri J, Ghosh DK, Mandal A (1997) Mercury and organomercurial resistance in bacteria isolated from freshwater fish of wetland fisheries around Calcutta. Environ Pollut 97:71–78
Schaefer JK, Letowski J, Barkay T (2002) mer mediated resistance and volatilization of Hg(II) under anaerobic conditions. Geomicrobiol J 19:87–102
Schaefer JK, Yagi J, Reinfelder JR, Cardona T, Ellickson KM, Tel-Or S, Barkay T (2004) Role of the bacterial organomercury lyase (MerB) in controlling methylmercury accumulation in mercury-contaminated natural waters. Environ Sci Technol 38:4304–4311
Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P (2004) Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol 186:427–437
Schelert J, Drozda M, Dixit V, Dillman A, Blum P (2006) Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J Bacteriol 188:7141–7150
Shrestha RA, Sillanpää M (2008) Influence of Eh/pH-Barriers on releasing/accumulation of manganese and iron at sediment-water interface. Res J Chem Environ 12:7–13
Shrestha R, Kafle B, Sillanpää M (2010) Water quality of Dhulikhel area, Nepal. Res J Chem Environ 14:36–38
Siciliano SD, O’Driscoll NJ, Lean DRS (2002) Microbial reduction and oxidation of mercury in freshwater lakes. Environ Sci Technol 36:3064–3068
Sillanpää M (2009) Occurrence, interaction with heavy metals and behaviour of complexing agents in the environment: a review. Res J Chem Environ 13:99–103
Sillanpää M, Oikari A (1996) Assessing the impact of complexation by EDTA and DTPA on heavy metal toxicity using microtox bioassay. Chemosphere 32:1485–1497
Sillanpää M, Rämö J (2009) Metal analysis of pulp: ICP-AES, XRF and ISE methods and their on-line feasibility. Res J Chem Environ 13:63–67
Sillanpää M, Orama M, Rämö J, Oikari A (2001) The importance of ligand speciation in environmental research: a case study. Sci Tot Environ 267:23–31
Silver S, Phung LT (1996) Bacterial heavy metal resistance; new surprises. Ann Rev Microbiol 50:753–789
Sinha A, Khare SK (2010) Mercury bioaccumulation and simultaneous nanoparticle synthesis by Enterobacter sp. cells. Bioresour Technol 102:4281–4284
Smalla K, Haines AS, Jones K, Krögerrecklenfort E, Heuer H, Schloter M, Thomas CM (2006) Increased abundance of IncP-1beta plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1beta plasmids with a complex mer transposon as the sole accessory element. Appl Environ Microbiol 72(11):7253–7259
Soge OO, Beck NK, White TM, No DB, Robert M (2008) A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J Antimicrob Chemother 62:674–680
Sorvari J, Sillanpää M (1996) Influence of metal complex formation on heavy metal and free EDTA and DTPA acute toxicity determined by D. magna. Chemosphere 33:1119–1127
Sugio T, Komoda T, Okazaki Y, Takeda Y, Nakamura S, Takeuchi F (2010) Volatilization of metal mercury from organomercurial by highly mercury-resistant Acidithiobacillus ferrooxidans MON-1. Biosci Biotechnol Biochem 74:1007–1012
Summers AO, Sugarman LI (1974) Cell-free mercury(II)-reducing activity in a plasmid-bearing strain of Escherichia coli. J Bacteriol 119:242–249
Summers AO, Wireman J, Vimy MJ, Lorscheider FL, Marshall B, Levy SB, Bennett S, Billard L (1993) Mercury released from dental “silver” fillings provokes an increase in mercury—and antibiotic-resistant. Antimicrob Agents Chemother 37:825–834
Vanasse JL, Lefebvre M, Lello PD, Sygusch J, Omichinsk JG (2008) Crystal structures of the organomercurial lyase MerB in its free and mercury-bound forms. J Biol Chem 284:938–944
Vetriani C, Chew YS, Miller SM, Yagi J, Coombs J, Lutz RA, Barkay T (2005) Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl Environ Microbiol 71:220–226
Vilhunen SH, Sillanpää MET (2009) Ultraviolet light emitting diodes and hydrogen peroxide in the photodegradation of aqueous phenol. J Hazard Mater 161:1530–1534
Vilhunen S, Särkkä H, Sillanpää M (2009) Ultraviolet light emitting diodes in water disinfection. Environ Sci Pollut Res 16:439–442
Vilhunen S, Puton J, Virkutyte J, Sillanpää M (2011) Efficiency of hydroxyl radical formation and phenol decomposition by using UV light emitting diodes and H2O2. Environ Technol 32:865–872
Virkutyte J, Sillanpää M (2006) Chemical evaluation of potable water in eastern Qinghai province, China: human health aspects. Environ Int 32:80–86
Wang Y, Freedman Z, Lu-Irving P, Kaletsky R, Barkay R (2009) An initial characterization of the mercury resistance (mer) system of the thermophilic bacterium Thermus thermophilus HB27. FEMS Microbiol Ecol 67:118–129
Weber JH, Evans R, Jones SH, Hines ME (1998) Conversion of mercury(II) into mercury(0), monomethylmercury cation, and dimethylmercury in saltmarsh sediment slurries. Chemosphere 36:1669–1687
Wireman J, Liebert CA, Smith T, Summers AO (1997) Association of mercury resistance with antibiotic resistance in the gram-negative fecal bacteria of primates. Appl Environ Microbiol 63:4494–4503
Zeyaullah Md, Islam B, Arif Ali A (2010) Isolation, identification and PCR amplification of merA gene from highly mercury polluted Yamuna river. AJB 9:3510–3514
Conflict of interest
Authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Mathema, V.B., Thakuri, B.C. & Sillanpää, M. Bacterial mer operon-mediated detoxification of mercurial compounds: a short review. Arch Microbiol 193, 837–844 (2011). https://doi.org/10.1007/s00203-011-0751-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-011-0751-4