Abstract
Sparse convex clustering is to group observations and conduct variable selection simultaneously in the framework of convex clustering. Although a weighted \(L_1\) norm is usually employed for the regularization term in sparse convex clustering, its use increases the dependence on the data and reduces the estimation accuracy if the sample size is not sufficient. To tackle these problems, this paper proposes a Bayesian sparse convex clustering method based on the ideas of Bayesian lasso and global-local shrinkage priors. We introduce Gibbs sampling algorithms for our method using scale mixtures of normal distributions. The effectiveness of the proposed methods is shown in simulation studies and a real data analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cluster analysis is an unsupervised learning method aimed at assigning observations to several clusters so that similar individuals belong to the same group, and is widely used in various research fields as biology and genomics, as well as many other fields of science. Until now, many clustering methods have been proposed: hierarchical clustering, k-means clustering (Hartigan and Wong 1979), Gaussian mixture model (McLachlan et al. 2019), Bayesian nonparametric clustering (Frühwirth-Schnatter and Malsiner-Walli 2019; Wade and Ghahramani 2018), and Bayesian clustering based on the uncertainty (Chandra et al. 2020; Rigon et al. 2020). However, in general, these clustering methods are instable due to non-convex optimization.
Convex clustering proposed by Hocking et al. (2011) searches for the centers of all clusters simultaneously with allocating individuals to the clusters. Convex relaxation ensures that it achieves a unique global optimum regardless of the initial values. Estimates can be obtained by solving a regularization problem, which is similar to sparse regularization in regression analysis. However, convex clustering does not work well if the data contain a large amount of irrelevant features.
Sparse regularization is used to exclude irrelevant information. Wang et al. (2018) proposed sparse convex clustering to perform convex clustering and variable selection simultaneously. Sparse convex clustering estimates sparse models by using the \(L_1\) norm in addition to the regularization term of the convex clustering. Also, Wang et al. (2018) used the \(L_1\) norm for the convex clustering penalties, where the penalty was assumed to be different weights according to individual and feature. However, it was pointed out by Griffin and Brown (2011) that the penalty used in sparse convex clustering depends on the data, which may lead to model estimation accuracy degradation when the sample size is small.
Our proposed methods overcome the problem that penalties in sparse convex clustering depend heavily on weights. In particular, with these methods, even when the sample size is small, estimation is possible without depending on the weights. To propose a method, we first introduce a Bayesian formulation of sparse convex clustering, and then propose a Bayesian sparse convex clustering based on a global-local (GL) prior distribution. As the GL prior, we consider four types of distributions: a normal-exponential-gamma distribution (Griffin and Brown 2005), a normal-gamma distribution (Brown and Griffin 2010), a horseshoe distribution (Carvalho et al. 2010), and a Dirichlet–Laplace distribution (Bhattacharya et al. 2015). The Gibbs sampling algorithm for our proposed models is derived by using scale mixtures of normal distributions (Andrews and Mallows 1974). We note that although many other GL prior distributions have been proposed, we use representative prior distributions among them. Recently, many researchers have discussed their relationships: Bhadra et al. (2019), Cadonna et al. (2020), Van Erp et al. (2019), Bhadra et al. (2017), and Piironen and Vehtari (2017).
The rest of this paper is organized as follows. Section 2 focuses on the sparse convex clustering method. In Sect. 3, we propose a Bayesian formulation of sparse convex clustering. A Bayesian convex clustering method with GL shrinkage prior distributions is proposed in Sect. 4. The performances of the proposed methods are compared with those of the existing method by conducting a Monte Carlo simulation in Sect. 5 and a real data analysis in Sect. 6. Concluding remarks are given in Sect. 7.
2 Sparse convex clustering
In this section, we describe convex clustering. This is a convex relaxation of such clustering methods as k-means clustering and hierarchical clustering. The convexity overcomes the instability of conventional clustering methods. In addition, we describe sparse convex clustering which simultaneously clusters observations and performs variable selection.
Let \(X \in {\mathbb {R}}^{n\times p}\) be a data matrix with n observations and p variables, and \({\varvec{x}}_i\ ( i=1,2,\ldots ,n)\) be the i-th row of X. Convex clustering for these n observations is formulated as the following minimization problem using an \(n \times p\) feature matrix \( A=({\varvec{a}}_1,\ldots ,{\varvec{a}}_n)^T \):
where \({\varvec{a}}_i\) is a p-dimensional vector corresponding to \({\varvec{x}}_i\), \(\Vert \cdot \Vert _q\) is the \(L_q\) norm of a vector, and \(\gamma \ (\ge 0)\) is a regularization parameter. If \(\hat{{\varvec{a}}}_{i_1}=\hat{{\varvec{a}}}_{i_2}\) for the estimated value \(\hat{{\varvec{a}}}_i\), then the \(i_1\)-th individual and \(i_2\)-th individual belong to the same cluster. The \(\gamma \) controls the number rows of \({\hat{A}} = (\hat{{\varvec{a}}}_1, \ldots , \hat{{\varvec{a}}}_n)^T\) that are the same, which determines the estimated number of clusters. Both k-means clustering and hierarchical clustering are equivalent to considering the \(L_0\) norm for the second term in the problem (1), which becomes a non-convex optimization problem (Hocking et al. 2011). Convex clustering can be viewed as a convex relaxation of k-means clustering and hierarchical clustering. This convex relaxation guarantees that a unique global minimization is achieved.
Hocking et al. (2011) proposed using a cluster path to visualize the steps of clustering. A cluster path can be regarded as a continuous regularization path (Efron et al. 2004) of the optimal solution formed by changing \(\gamma \). Figure 1 shows the cluster path of two interlocking half-moons described in Sect. 5.1. A cluster path shows the relationship between values of the regularization parameter and estimates of the feature vectors. The estimates exist near the corresponding observations when the value of the regularization parameter is small, while the estimates concentrate on one point when the value is large. The characteristics of the data can be considered from the grouping order and positional relationship of the estimates.
A cluster path for two interlocking half-moons. The colored squares are 20 observations and the circles are convex clustering estimates for different regularization parameter values. Among the estimates of the same observation, the lines connect the estimates whose values of the regularization parameter are close
In conventional convex clustering, when irrelevant information is included in the data, the accuracy of estimating clusters tends to be low. Sparse convex clustering (Wang et al. 2018), on the other hand, is an effective method for such data, as irrelevant information can be eliminated using sparse estimation.
Sparse convex clustering considers the following optimization problem:
where \(\gamma _1\ (\ge 0)\) and \(\gamma _2\ (\ge 0)\) are regularization parameters, \(w_{i_1,i_2}\ (\ge 0)\) and \(u_j\ (\ge 0)\) are weights, \(q \in \{1, 2, \infty \}\), \({\mathcal {E}}=\{(i_1,i_2); w_{i_1,i_2} \ne 0, i_1<i_2\}\), and \({\varvec{a}}_{\cdot j} = (a_{1j},\ldots ,a_{nj})^T\) is a column vector of the feature matrix A. The third term imposes a penalty similar to group lasso (Yuan and Lin 2006) and has the effect that \(\Vert \hat{{\varvec{a}}}_{\cdot j}\Vert _1 = 0\). When \(\Vert \hat{{\varvec{a}}}_{\cdot j}\Vert _1 = 0\), the j-th column of X is removed from the model, which is variable selection. \(\gamma _1\) and \(w_{i_1,i_2}\) adjust the cluster size, whereas \(\gamma _2\) and \(u_j\) adjust the number of features. The weight \(w_{i_1,i_2}\) plays an important role in imposing a penalty that is adaptive to the features. Wang et al. (2018) used the following weight parameter:
where \(\iota _{i_1, i_2}^{m}\) equals 1 if the observation \({\varvec{x}}_{i_1}\) is included among the m nearest neighbors of the observation \({\varvec{x}}_{i_2}\), and is 0 otherwise. This choice of weights works well for a wide range of \(\phi \) when m is small. In our numerical studies, m is fixed at 5 and \(\phi \) is fixed at 0.5, as in Wang et al. (2018).
Similar to the adaptive lasso (Zou 2006) in a regression problem, the penalty for sparse convex clustering can be adjusted flexibly by using weight parameters. However, it was shown by Griffin and Brown (2011) that such penalties are strongly dependent on the data. In particular, the accuracy of model estimation may be low due to the data, such as because the number of samples is small.
We remark that Yau and Holmes (2011) and Malsiner-Walli et al. (2016) have proposed Gaussian mixture models for clustering and variable selection simultaneously. While sparse convex clustering is convex optimization, these methods are non-convex optimization.
3 Bayesian formulation of sparse convex clustering
By extending sparse convex clustering to a Bayesian formulation, we may use the entire posterior distribution to provide a probabilistic measure of uncertainty.
3.1 Bayesian sparse convex clustering
In this section, we reformulate sparse convex clustering as a Bayesian approach. Similar to Bayesian lasso (Park and Casella 2008), which extends lasso to a Bayesian formulation, we regard regularized maximum likelihood estimates as MAP estimates.
We consider the following model:
where \({\varvec{\varepsilon }}\) is a p-dimensional error vector distributed as \(\text{ N}_p({\varvec{0}}_p, \sigma ^2I_p)\), \({\varvec{a}}\) is a feature vector, and \(\sigma ^2\ (>0)\) is a variance parameter. Then, the likelihood function is given by
Next, we specify the prior distribution of feature matrix A as
where \(\lambda _1\ (>0),\ w_{i_1,i_2}\ (>0),\ \lambda _2\ (>0),\ u_j\ (>0)\) are hyperparameters, \({\mathcal {E}} = \{(i_1,i_2): 1 \le i_1<i_2\le n\}\), and \(\# {\mathcal {E}}\) is the number of elements in \({\mathcal {E}}\). Note that \(\lambda _1\) and \(\lambda _2\) correspond to \(\gamma _1\) and \(\gamma _2\) in (2), respectively. This prior distribution is an extension of Bayesian group lasso in linear regression models (Xu and Ghosh 2015). The estimate of a specific sparse convex clustering corresponds to the MAP estimate in the following joint posterior distribution:
where \(\pi (\sigma ^2)\) is the non-informative scale-invariant prior \(\pi (\sigma ^2) \propto 1/\sigma ^2\) or inverse-gamma prior \(\pi (\sigma ^2)=\text{ IG }(\nu _0/2,\eta _0/2)\). An inverse-gamma probability density function is given by
where \(\nu \ (>0)\) is a shape parameter, \(\eta \ (>0)\) is a scale parameter, and \(\varGamma (\cdot )\) is the gamma function. Note that this prior distribution (4) has unimodality: see “Appendix A”.
We obtain estimates of each parameter by applying the MCMC algorithm with Gibbs sampling. Therefore, it is necessary to derive the full conditional distribution for each parameter. Because it is difficult to derive full conditional distributions from (4), we derive a hierarchical representation of the prior distribution. From this relationship, we assume the following priors:
These priors enable us to carry out Bayesian estimation using Gibbs sampling. The details of the hierarchical representation of the prior distribution (4) and the sampling procedure are described in “Appendix B.1”.
3.2 MAP estimate by weighted posterior means
In Bayesian sparse modeling, an unweighted posterior mean is often used as a substitute for MAP estimates, but the accuracy is not high and sometimes it is far from the MAP estimates. As a result, we introduce the weighted posterior mean in this section.
We define a vector \({\varvec{\theta }}\) containing all the parameters as follows:
where \({\varvec{\tau }}_i=(\tau _{i1}, \ldots , \tau _{in})\) and \(\widetilde{{\varvec{\tau }}}=({\widetilde{\tau }}_{1}, \ldots , {\widetilde{\tau }}_{p})\). For example, \({\varvec{\theta }}_1={\varvec{a}}_1\) and \({\varvec{\theta }}_{n+1}={\varvec{\tau }}_1\). In addition, we assume the parameter vector corresponding to the b-th MCMC sample is \({\varvec{\theta }}^{(b)}=({\varvec{\theta }}_1^{(b)},\ldots ,{\varvec{\theta }}_{2n+2}^{(b)}) \), where the range of b is from 1 to B.
We introduce weights corresponding to the b-th MCMC sample as follows:
where \(L(X|{\varvec{\theta }})\) is the likelihood function, \(\pi ({\varvec{\theta }})\) is the prior,
and \(\hat{{\varvec{\theta }}}_{l'}\) is an estimate of \({\varvec{\theta }}_{l'}\). It can be seen that this weight corresponds to the value of the posterior probability according to Bayes’ theorem. This weight was also used in the sparsified algorithm proposed by Shimamura et al. (2019).
Using this weight, we obtain the posterior average as follows:
where \(w_{({\varvec{\theta }}_l, b)}={\widetilde{w}}_{({\varvec{\theta }}_l, b)}/\sum _{b'=1}^B{\widetilde{w}}_{({\varvec{\theta }}_l, b')}\). Therefore, we adopt \(\hat{{\varvec{\theta }}}_l\) as an estimate of \({\varvec{\theta }}_l\). The performance of this estimate is examined by numerical studies in Sect. 5.1.
4 Bayesian sparse convex clustering via global-local (GL) shrinkage priors
Polson and Scott (2010) proposed a GL shrinkage prior distribution. Generally speaking, when we use the Laplace prior distribution, it is necessary to pay attention to how to handle contraction for irrelevant parameters and robustness against relevant parameters. The important features of the GL shrinkage prior distribution are that it has a peak at the origin and heavy tails. These features make it possible to handle shrinkage of all variables, and the individual variables shrinkage estimated to be zero. Therefore, irrelevant parameters are sparsified, and relevant ones are robustly estimated. The penalty for sparse convex clustering has similar characteristics. Specifically, it is weighted on individual and feature quantities. This weighted penalty is one of the key factors for improving accuracy. However, this penalty has the problem that it is highly dependent on the data. By using the GL prior distribution, it is possible to properly control this the dependency by using the Bayesian approach.
Polson and Scott (2010) formulated the GL scale mixtures of normal distributions for vector \({\varvec{a}} = (a_1,\ldots ,a_p)\) as follows:
Each \(\tau _j^2\ (>0)\) is called a local shrinkage parameter and \(\nu \ (>0)\) is called a global shrinkage parameter. This leads to efficient Gibbs sampling based on block updating of parameters.
We need to specify the priors \(\pi (\tau _j^2)\) and \(\pi (\nu ^2)\). In the next subsections, we provide some concrete formulae for \(\pi (\tau _j^2)\) and \(\pi (\nu ^2)\).
4.1 NEG prior distribution
Griffin and Brown (2005) proposed using an NEG distribution as an alternative to a Laplace distribution for the prior distribution of regression coefficients. By using an NEG distribution, we can perform more flexible sparse modeling than with a Laplace distribution.
The NEG density function is given by
where \(\kappa = \displaystyle (2^\lambda \lambda )/(\gamma \sqrt{\pi }){\Gamma }(\lambda +1/2)\) is a normalization constant, \(D_{-2\lambda -1}\) is a parabolic cylinder function, and \(\lambda \ (>0)\) and \(\gamma \ (>0)\) are hyperparameters that control the sparsity of \(\theta \). The parabolic cylinder function is a solution of a second-order linear ordinary differential equation and its integral representation is given by
The NEG density function can be expressed as hierarchical representation
where \(\text{ Exp }(\cdot |\mu )\) is the exponential distribution and \(\text{ Ga }(\cdot |k, \lambda )\) is a gamma distribution. Therefore, the prior distribution of each parameter is as follows:
Using the NEG distribution on the feature matrix A, we propose the following prior:
By using the hierarchical representation, we can develop a Gibbs sampling algorithm for Bayesian sparse convex clustering with NEG prior distributions. The details of the hierarchical representation and the algorithm are given in “Appendix B.2”.
4.2 NG prior distribution
Brown and Griffin (2010) proposed an NG distribution as follows:
where \(\lambda \ (>0)\) and \(\gamma \ (>0)\) are hyperparameters that control the sparsity of \(\theta \). The NG prior is a generalization of a Laplace distribution and has been used successfully in many applications. For example, Malsiner-Walli et al. (2016) used the NG prior in a sparse finite mixture model.
Using the NG distribution on the feature matrix A, we propose the following prior:
Note that this prior distribution consists of the NG distribution and the NEG distribution. The prior distribution can also be constructed using only the NG distribution. However, as a result of the numerical experiment, it did not work well. Therefore, we adopt the NEG distribution for the prior that induces variable selection.
By using the hierarchical representation, we can develop a Gibbs sampling algorithm for Bayesian sparse convex clustering with NG prior distributions. The details of the hierarchical representation and the algorithm are given in “Appendix B.3”.
4.3 Horseshoe prior distribution
The horseshoe density function (Carvalho et al. 2010) is given by
The prior distribution of each parameter is as follows:
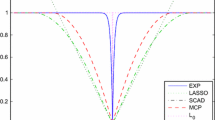
Here \(\nu \ (>0)\) is a hyperparameter that controls the sparsity of the \(\theta _j\)’s, and \(\text{ C}^+(x_0, \gamma )\) is the half Cauchy distribution on the positive reals, where \(x_0\) is a location parameter and \(\gamma \) is a scale parameter. A smaller value of hyperparameter \(\nu \) corresponds to a higher, number of parameters \(\{\theta _j\}\) being estimated to be zero.
Using the horseshoe distribution on the feature matrix A, we propose the following prior:
where \({\varvec{\mathrm {a}}} = \left( {\Vert {\varvec{a}}_{i_1}-{\varvec{a}}_{i_2}\Vert _2;\ (i_1,i_2)\in {\mathcal {E}}} \right) \). Similar with reasons as in the prior (7), this prior distribution consists of the horseshoe distribution and the NEG distribution.
By using the hierarchical representation, we can develop a Gibbs sampling algorithm for Bayesian sparse convex clustering with horseshoe prior distributions. The details of the hierarchical representation and the algorithm are given in “Appendix B.4”. In our proposed method, we used \(\nu _1\) as a hyperparameter, while the approach based on Makalic and Schmidt (2015) assumed a prior distribution for \(\nu _1\). We compared both approaches through numerical experiments, but the approach by Makalic and Schmidt (2015) does not work well.
4.4 Dirichlet–Laplace prior distribution
The Dirichlet–Laplace prior was proposed to provide simple sufficient conditions for posterior consistency (Bhattacharya et al. 2015). It is known that a Bayesian regression model with this prior distribution has asymptotic posterior consistency with respect to variable selection. Also, we can obtain joint posterior distributions for a Bayesian regression model when we employ this prior. The latter is advantageous because most prior distributions induce a marginal posterior distribution rather than a joint posterior distribution, which has more information in general.
The Dirichlet–Laplace density function is given by
where \({\varvec{\tau }} = (\tau _1,\ldots ,\tau _p)^T\). The prior distribution of each parameter is
where \(\alpha \ (>0)\) is a hyperparameter that controls the sparsity of the \(\theta _j\)’s and \(\text{ Dir }(\alpha ,\ldots ,\alpha )\) is a Dirichlet distribution. Random variables of the Dirichlet distribution sum to one, and have mean \(\text{ E }[\tau _j]=1/p\) and variance \(\text{ Var }(\tau _j) = (p-1)/\{p^2(p\alpha +1)\}\). When \(\alpha \) is small, most of the parameters \(\{\tau _j\}\) are close to zero, whereas the remaining parameters are close to one. If \(\{\tau _j\}\) is close to zero, \(\{\theta _j\}\) is also close to zero.
Using the Dirichlet–Laplace distribution on the feature matrix A, we propose the following prior:
Similar with reasons as in the prior (7), this prior distribution consists of the Dirichlet-Laplace distribution and the NEG distribution.
By using the hierarchical representation, we can develop a Gibbs sampling algorithm for Bayesian sparse convex clustering with Dirichlet–Laplace prior distributions. The details of the hierarchical representation and the algorithm are given in “Appendix B.5”.
5 Artificial data analysis
In this section, we describe numerical studies to evaluate the performance of the proposed methods using artificial data. First, clustering performance was evaluated by an illustrative example that includes no irrelevant features. Next, we evaluated the accuracy of the sparsity by performing simulations using data containing irrelevant features.
5.1 Illustrative example
We demonstrated our proposed methods with artificial data. The data were generated according to two interlocking half-moons with \(n=50\) observations, \(K=2\) clusters, and \(p=2\) features. Figure 2 shows one example of two interlocking half-moons. In this setting, we did not perform sparse estimation. The cluster formation was considered by comparing the cluster paths of each method.
For each generated dataset, the estimates were obtained by using 50,000 iterations of a Gibbs sampler. Candidates of the hyperparameters were set based on
for \(i=1,\ldots ,m\). For the hyperparameter \(\lambda \) in Bayesian convex clustering with a Laplace prior distribution (Bscvc), we set \(m = 50\), \(\lambda _{\min } = 0.05\), and \(\lambda _{\max } = 90.0\). In Bayesian convex clustering with an NEG prior distribution (NEG), we had hyperparameters \(\lambda _1\) and \(\gamma _1\). For hyperparameter \(\lambda _1\), we set \(m = 30\), \(\lambda _{\min } = 0.0001\), and \(\lambda _{\max } = 2.75\). For hyperparameter \(\gamma _1\), we set \(m = 2\), \(\lambda _{\min } = 0.4\), and \(\lambda _{\max } = 0.5\). The weighted posterior means introduced in Sect. 3.2 were used for Bscvc, NEG, NG, HS, and DL estimates.
Figure 3 shows the results. The overall outline of cluster formation is the same for the all methods. The order in which the samples form clusters is also the same. If the distance between estimated feature values of different clusters does not decrease, the accuracy of cluster estimation will improve in convex clustering. However, the distances between all features are small due to the effect of sparse regularization. Scvc used weights to bring only features belonging to the same cluster closer. NEG, NG, HS, and DL used GL priors instead of weights. For example, in the cluster path in Fig. 3b, the estimated feature values are merged at a position further from the origin than other methods. This can be seen especially in the upper right and lower left of the figure. This result shows that the close feature values were merged while the distances between the distant feature values were maintained. This is a factor that improves the accuracy of NEG’s clustering estimation.
5.2 Simulation studies
We demonstrated our proposed methods with artificial data including irrelevant features. First, we considered five settings. Each data were generated according to two interlocking half-moons with \(n=10\), 20, 50, 100 observations, \(K=2\) clusters, and \(p=20\), 40 features. The features consisted \(p-2\) irrelevant features and 2 relevant features. The irrelevant features were independently generated from \(\text{ N }(0, 0.5^2)\). We considered six methods: sparse convex clustering (Scvc), Bscvc, NEG, NG, HS, and DL.
As the estimation accuracy, we used the RAND index, which is a measure of correctness of cluster estimation. The RAND index ranges between 0 and 1, with a higher value indicating better performance. The RAND index is given by
where
Here \({\mathcal {C}}^*=\{{\mathcal {C}}_1^*,\ldots ,{\mathcal {C}}_r^*\}\) is the true set of clusters and \(\widetilde{{\mathcal {C}}}=\{\widetilde{{\mathcal {C}}}_1,\ldots ,\widetilde{{\mathcal {C}}}_s\}\) is the estimated set of clusters. In addition, we used the true negative rate (TNR) and the true positive rate (TPR) for the accuracy of sparse estimation:
where, \(\{{\varvec{a}}_j^*|j=1,\ldots ,p\}\) are the true feature vectors and \(\{\hat{{\varvec{a}}}_j|j=1,\ldots ,p\}\) are the estimated feature vectors. The dataset was generated 50 times. We computed the mean and standard deviation of RAND, TNR, and TPR from the 50 repetitions. The settings of the iteration count and the hyperparameter candidate were the same as given in Sect. 5.1. To ensure fair comparisons, we used the results with hyperparameters that maximize the RAND index.
The simulation results were summarized in Table 1. Scvc provided the lower RANDs and TNRs than other methods in all settings. TPR was competitive among the all methods. Except for Scvc, NEG, NG, HS, and DL were better than Bscvc in terms of RAND in almost all settings. From these experiments, we observed that the Bayesian convex clustering methods were superior to the conventional convex clustering method. In addition, the Bayesian methods based on the GL priors relatively produced the higher RANDs than those based on the Laplace prior.
6 Application
We applied our proposed methods to a real dataset: the LIBRAS movement data from the Machine Learning Repository (Lichman 2013). The LIBRAS movement dataset has 15 classes. Each class was divided by type of hand movement. There are 24 observations in each class, and each observation has 90 features consisting of hand movement coordinates. In this numerical experiment, 5 classes were selected from among the 15 classes that were the same classes as those selected by Wang et al. (2018). Accuracies of each method were evaluated using the RAND index, the estimated number of clusters, and the number of selected features. This is the same procedure as reported in Wang et al. (2018). As in Sect. 5.2, we used the results with hyperparameters that maximize the RAND index for comparisons.
The results are summarized in Table 2. All the methods provided the same RAND. Although the true number of clusters is six, the inherent number of clusters might be five because the corresponding RAND is highest among all methods. Scvc, Bscvc, and NG selected all features, while NEG, HS, and DL selected some of features. In other words, NEG, HS, and DL could be sparsified without degrading the accuracy of cluster estimation. In addition, we performed principal component analysis to this real dataset. Figure 4 displays the plot of the first principal component against the second principal component. The numbers in Fig. 4a stand for the true groups. The numbers and the colors in Fig. 4b stand for the true groups and estimated groups, respectively. From Fig. 4, we observe that the groups 4 and 6 have the same color, which means that our proposed method could not capture the characteristic between the numbers 4 and 6.
7 Conclusion
We proposed a Bayesian formulation of the sparse convex clustering. Using the GL shrinkage prior distribution, we constructed a Bayesian model for various data with more flexible constraints than ordinary \(L_1\)-type convex clustering. We overcame the problem that sparse convex clustering depends on weights in the regularization term. Furthermore, we proposed a weighted posterior mean based on a posteriori probability to provide more accurate MAP estimation.
For the application described in Sect. 6, the computational time with our proposed methods was about 20 minutes for each hyperparameter. Until now, it is difficult to compute our MCMC methods when the dimension is high. Recently, the variational Bayesian method has been received attention [e.g., Ray and Szabó (2020) and Wang and Blei (2019)] as an alternative to MCMC. In addition, Johndrow et al. (2020) proposes to speed up MCMC by improving the conventional sampling method. We would like to work on reducing the computational cost to expand the field of applicability with the techniques of these studies. In our numerical experiment, the hyperparameters with the best accuracy were selected using the same method as reported in Wang et al. (2018). It would also be interesting to develop information criteria for selecting the hyperparameters. We leave these topics as future work.
References
Andrews DF, Mallows CL (1974) Scale mixtures of normal distributions. J R Stat Soc B 36(1):99–102
Bhadra A, Datta J, Polson NG, Willard B (2019) Lasso meets horseshoe: a survey. Stat Sci 34(3):405–427
Bhadra A, Datta J, Polson NG, Willard B et al (2017) The horseshoe+ estimator of ultra-sparse signals. Bayesian Analysis 12(4):1105–1131
Bhattacharya A, Pati D, Pillai NS, Dunson DB (2015) Dirichlet–Laplace priors for optimal shrinkage. J Am Stat Assoc 110(512):1479–1490
Brown PJ, Griffin JE (2010) Inference with normal-gamma prior distributions in regression problems. Bayesian Anal 5(1):171–188
Cadonna A, Frühwirth-Schnatter S, Knaus P (2020) Triple the gamma—a unifying shrinkage prior for variance and variable selection in sparse state space and TVP models. Econometrics 8(2):20
Carvalho CM, Polson NG, Scott JG (2010) The horseshoe estimator for sparse signals. Biometrika 97(2):465–480
Chandra NK, Canale A, Dunson DB (2020) Bayesian clustering of high-dimensional data. arXiv preprint arXiv:2006.02700
Efron B, Hastie T, Johnstone I, Tibshirani R (2004) Least angle regression. Ann Stat 32(2):407–499
Frühwirth-Schnatter S, Malsiner-Walli G (2019) From here to infinity: sparse finite versus Dirichlet process mixtures in model-based clustering. Adv Data Anal Classif 13(1):33–64
Griffin JE, Brown PJ (2005) Alternative prior distributions for variable selection with very many more variables than observations. University of Kent Technical Report
Griffin JE, Brown PJ (2011) Bayesian hyper-lassos with non-convex penalization. Aust N Z J Stat 53(4):423–442
Hartigan JA, Wong MA (1979) A k-means clustering algorithm. J R Stat Soc Ser C (Appl Stat) 28(1):100–108
Hocking TD, Joulin A, Bach F, Vert J-P (2011) Clusterpath : an algorithm for clustering using convex fusion penalties. In: Proceedings of the 28th international conference on machine learning (ICML)
Johndrow JE, Orenstein P, Bhattacharya A (2020) Scalable approximate MCMC algorithms for the horseshoe prior. J Mach Learn Res 21(73):1–61
Lichman M (2013) UCI machine learning repository, 2013. http://archive.ics.uci.edu/ml
Makalic E, Schmidt DF (2015) A simple sampler for the horseshoe estimator. IEEE Signal Process Lett 23(1):179–182
Malsiner-Walli G, Frühwirth-Schnatter S, Grün B (2016) Model-based clustering based on sparse finite Gaussian mixtures. Stat Comput 26(1–2):303–324
McLachlan GJ, Lee SX, Rathnayake SI (2019) Finite mixture models. Annu Rev Stat Appl 6:355–378
Park T, Casella G (2008) The Bayesian Lasso. J Am Stat Assoc 103:681–686
Piironen J, Vehtari A et al (2017) Sparsity information and regularization in the horseshoe and other shrinkage priors. Electron J Stat 11(2):5018–5051
Polson NG, Scott JG (2010) Shrink globally, act locally: Sparse Bayesian regularization and prediction. Bayesian Stat 9:501–538
Ray K, Szabó B (2020) Variational Bayes for high-dimensional linear regression with sparse priors. J Am Stat Assoc 1–31
Rigon T, Herring AH, Dunson DB (2020) A generalized Bayes framework for probabilistic clustering. arXiv preprint arXiv:2006.05451
Shimamura K, Ueki M, Kawano S, Konishi S (2019) Bayesian generalized fused lasso modeling via neg distribution. Commun Stat Theory Methods 48(16):4132–4153
Van Erp S, Oberski DL, Mulder J (2019) Shrinkage priors for Bayesian penalized regression. J Math Psychol 89:31–50
Wade S, Ghahramani Z et al (2018) Bayesian cluster analysis: point estimation and credible balls (with discussion). Bayesian Anal 13(2):559–626
Wang B, Zhang Y, Sun WW, Fang Y (2018) Sparse convex clustering. J Comput Graph Stat 27(2):393–403
Wang Y, Blei DM (2019) Frequentist consistency of variational Bayes. J Am Stat Assoc 114(527):1147–1161
Xu X, Ghosh M (2015) Bayesian variable selection and estimation for group lasso. Bayesian Anal 10(4):909–936
Yau C, Holmes C (2011) Hierarchical Bayesian nonparametric mixture models for clustering with variable relevance determination. Bayesian Anal (Online) 6(2):329
Yuan M, Lin Y (2006) Model selection and estimation in regression with grouped variables. J R Stat Soc B 68(1):49–67
Zou H (2006) The adaptive lasso and its oracle properties. J Am Stat Assoc 101(476):1418–1429
Acknowledgements
The authors thank the reviewers for their helpful comments and constructive suggestions. S. K. was supported by JSPS KAKENHI Grant Numbers JP19K11854 and JP20H02227 and MEXT KAKENHI Grant Numbers JP16H06429, JP16K21723, and JP16H06430. Super computing resources were provided by Human Genome Center (the Univ. of Tokyo).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Unimodality of joint posterior distribution
In Bayesian modeling, theoretical and computational problems arise when there exist multiple posterior modes. Theoretically, it is doubtful whether a single posterior mean, median, or mode will appropriately summarize the bimodal posterior distribution. The convergence speed of Gibbs sampling presents a computational problem, in that, although it is possible to perform Gibbs sampling, the convergence is too slow in practice.
Park and Casella (2008) showed that the joint posterior distribution has a single peak in Lasso-type Bayes sparse modeling. We demonstrate that the joint posterior distribution of (4) is unimodal. Specifically, similar to Park and Casella (2008), we use a continuous transformation with a continuous inverse to show the unimodality of the logarithmic concave density.
The logarithm of the posterior (4) is
Consider the transformation defined by
which is continuous when \(0< \sigma ^2 < \infty \). We define \(\varPhi =({\varvec{\phi }}_1,\ldots ,{\varvec{\phi }}_n)^T=( {\varvec{\phi }}_{\cdot 1},\ldots ,{\varvec{\phi }}_{\cdot p})\). The log posterior (9) is transformed by performing variable conversion in the form
The second and fifth terms are clearly concave in \((\varPhi ,\rho )\), and the third and fourth terms are a concave surface in \((\varPhi ,\rho )\). Therefore, if \(\log \pi (\cdot )\), which is the logarithm of the prior for \(\sigma ^2\), is concave, then (10) is concave. Assuming a prior distribution, such as the inverse gamma distribution (5) for \(\sigma ^2\), \(\log \pi (\cdot )\) is a concave function. Therefore, the entire log posterior distribution is concave.
Appendix B: Formulation of Gibbs sampling
This appendix introduces a specific Gibbs sampling method for a Bayesian sparse convex clustering.
1.1 B.1 Bayesian sparse convex clustering
The prior distribution \(\pi (A|\sigma ^2)\) is rewritten as follows:
This representation is based on the following hierarchical representation of the Laplace distribution:
For details, we refer the reader to Andrews and Mallows (1974).
The prior distribution is transformed as follows:
The full conditional distribution is obtained as follows:
where \(\text{ IGauss }(x|\mu , \lambda )\) denotes the inverse-Gaussian distribution with density function
and
1.2 B.2 Bayesian NEG sparse convex clustering
By using the hierarchical representation of the NEG distribution, the prior distribution \(\pi (A|\sigma ^2)\) is decomposed into
This result allows us to develop a Gibbs sampling algorithm for Bayesian sparse convex clustering with the NEG prior distribution.
The prior distribution is transformed as follows:
The full conditional distribution is obtained as follows:
where
1.3 B.3 Bayesian NG sparse convex clustering
By using the hierarchical representation of the NG distribution, the prior distribution \(\pi (A|\sigma ^2)\) is decomposed into
This result allows us to develop a Gibbs sampling algorithm for Bayesian sparse convex clustering with the NG prior distribution.
The prior distribution is transformed as follows:
The full conditional distribution is obtained as follows:
where \(\text{ giG }\left( x|\chi ,\rho ,\lambda \right) \) is generalized inverse Gaussian
and
1.4 B.4 Bayesian horseshoe sparse convex clustering
By using the hierarchical representation of the horseshoe distribution, the prior distribution \(\pi (A|\sigma ^2)\) is obtained as follows:
The prior distribution is transformed as follows:
The full conditional distribution is obtained as follows:
where
1.5 B.5 Bayesian Dirichlet–Laplace sparse convex clustering
By using a hierarchical representation of the Dirichlet–Laplace distribution, the prior distribution \(\pi (A|\sigma ^2)\) is obtained as follows:
The prior distribution is transformed as follows:
The full conditional distribution is obtained as follows:
where
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shimamura, K., Kawano, S. Bayesian sparse convex clustering via global-local shrinkage priors. Comput Stat 36, 2671–2699 (2021). https://doi.org/10.1007/s00180-021-01101-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00180-021-01101-7