Abstract
Purpose
Venoarterial extracorporeal membrane oxygenation (VA-ECMO) is a complex and high-risk life support modality used in severe cardiorespiratory failure. ECMO survival scores are used clinically for patient prognostication and outcomes risk adjustment. This study aims to create the first artificial intelligence (AI)-driven ECMO survival score to predict in-hospital mortality based on a large international patient cohort.
Methods
A deep neural network, ECMO Predictive Algorithm (ECMO PAL) was trained on a retrospective cohort of 18,167 patients from the international Extracorporeal Life Support Organisation (ELSO) registry (2017–2020), and performance was measured using fivefold cross-validation. External validation was performed on all adult registry patients from 2021 (N = 5015) and compared against existing prognostication scores: SAVE, Modified SAVE, and ECMO ACCEPTS for predicting in-hospital mortality.
Results
Mean age was 56.8 ± 15.1 years, with 66.7% of patients being male and 50.2% having a pre-ECMO cardiac arrest. Cross-validation demonstrated an inhospital mortality sensitivity and precision of 82.1 ± 0.2% and 77.6 ± 0.2%, respectively. Validation accuracy was only 2.8% lower than training accuracy, reducing from 75.5% to 72.7% [99% confidence interval (CI) 71.1–74.3%]. ECMO PAL accuracy outperformed the ECMO ACCEPTS (54.7%), SAVE (61.1%), and Modified SAVE (62%) scores.
Conclusions
ECMO PAL is the first AI-powered ECMO survival score trained and validated on large international patient cohorts. ECMO PAL demonstrated high generalisability across ECMO regions and outperformed existing, widely used scores. Beyond ECMO, this study highlights how large international registry data can be leveraged for AI prognostication for complex critical care therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Extracorporeal membrane oxygenation predictive algorithm (ECMO PAL) is the first artificial intelligence (AI)-powered ECMO survival score trained and validated on a large international patient cohort. This work highlights how large international registry data can be leveraged for AI prognostication for complex critical care therapies. |
Introduction
Venoarterial extracorporeal membrane oxygenation (VA-ECMO) is an artificial heart–lung device used to support critically ill patients with cardiac and respiratory failure. Globally, VA-ECMO is increasingly used to support a wide variety of patients with numerous disease aetiologies [1]. VA-ECMO is among the most complex and expensive intensive care unit (ICU) therapies, costing from USD 42,000 to over USD 535,000 per run, requiring significant and often prolonged intensive care resources to maintain [2]. Due to the complexity of patients receiving VA-ECMO, the associated costs, and the high incidence of serious ECMO-related complications, appropriate patient selection and prognostication is crucial [3].
VA-ECMO outcome scores have been previously developed and used extensively for risk adjustment, patient prognostication, and quality control across time and centres [4,5,6,7,8,9,10,11,12]. Previously published scores have typically focused on ECMO outcomes for specific patient cohorts, such as cardiac arrest [6], cardiogenic shock [7,8,9,10], myocardial infarct [11], and post-coronary artery bypass graft [12]. The majority of these scores have been derived from patient data from a single centre or region, and thus their global generalisability may be limited. Furthermore, these scores have been derived using traditional statistical methods, typically univariate and multivariate analysis, with limited data-fitting capacity for complex questions, such as ECMO outcomes.
Artificial intelligence (AI) and machine learning (ML) offer an advanced alternative to traditional statistical methods and can be applied to large data cohorts with expansive variable sets, exposing complex interactions and patterns otherwise overlooked by traditional statistical approaches [13, 14].
This study aimed to leverage a large international patient cohort to develop and validate an AI-driven tool for predicting in-hospital mortality of VA-ECMO. The tool was derived entirely on pre-ECMO variables, allowing for mortality prediction immediately after ECMO initiation. AI-powered prognostication tools could improve ECMO centre quality control and benchmarking and with sufficient accuracy and demonstrated benefit, facilitate data-driven decisions about patient management.
Methods
These methods outline the full study protocol and adhere to the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD), Standard for Reporting of Diagnostic Accuracy Studies (STARD), and Minimum Information for Medical AI Reporting (MINIMAR) guidelines [15,16,17]. This research was approved by the Monash University Human Research Ethics Committee (#24863).
Data and inclusion
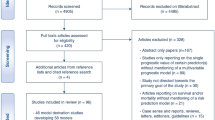
Data were obtained retrospectively from patients enrolled in the Extracorporeal Life Support Organisation (ELSO) registry [18]. The ELSO Registry collects data on the admission, initiation, maintenance, complications, and outcomes of ECMO patients worldwide. ELSO data are collected from 543 ECMO centres globally, primarily in the intensive care setting and other settings, such as the operating theatre or emergency department. The patient cohort comprised all adult patients (≥ 18 years) who received venoarterial ECMO between the 15th of January, 2017, and the 31st of December, 2020. This epoch was chosen to coincide with the latest major update to the ELSO registry reporting variables (January, 2017). The resulting cohort was a continuous set of 22,945 patients from 474 participating ECMO centres.
In the case of patients with multiple entries or a previous history of ECMO, only the first ECMO run was included. Three exclusion criteria were defined in consultation with senior ECMO intensivists; (1) patients indicated as still on ECMO whose outcome was unknown at the time of reporting (predominantly transferred to a non-ELSO participating centre and lost to follow-up), (2) presence of a durable ventricular assist device or total artificial heart, and (3) patient transported to reporting hospital on ECMO. Following exclusion, 18,167 patients remained for analysis (Fig. 1).
Exclusion criteria for training data were: patients still listed as receiving extracorporeal membrane oxygenation (ECMO), patients who had previously had ECMO, patients with a durable ventricular assist device (VAD) or total artificial heart (TAH), and patients transported to the reporting hospital on ECMO
Data pre-processing
All pre-ECMO registry variables were considered for analysis. Pre-ECMO variables were defined as the closest reading before ECMO initiation and collected no more than 6 h before ECMO initiation [18]. Data pre-processing consisted of deterministic manual data cleaning, data clustering, imputation, and feature scaling. Manual cleaning involved grouping ICD-10 diagnosis codes [19] into the following indications for VA-ECMO: acute myocardial infarction, arrhythmia, cardiomyopathy, myocarditis, post or perioperative support, pulmonary embolism, refractory cardiac arrest, sepsis, poisoning or toxins, any respiratory indication, and other cardiac or circulatory compromise (Supplementary Materials 1). These categories were chosen based on VA-ECMO indications found in the literature and other survival scores [20,21,22], with clustering performed independently by two clinicians and finalised through negotiation (MS and AD, > 99.99% independent agreement). In cases where the hospital admission diagnosis was given instead of the VA-ECMO indication, a deterministic method was used to derive VA-ECMO indication, combining additional patient data, such as surgical procedures and pre-ECMO support (Data Repository 1).
Data clustering was performed independently and negotiated by two of three clinicians (MS, AD, DP) and included the grouping of ICD-10 codes into comorbidities as identified in the Charlson Comorbidity Index [23] and Multipurpose Australian Comorbidity Scoring System (MACCS) [24]. Furthermore, clustering of Current Procedural Terminology (CPT [25]) codes was undertaken to identify pre-ECMO cardiothoracic surgery and other major pre-ECMO surgeries (Supplementary Materials 2). Pre-ECMO support was also considered, categorising drugs broadly by pharmacological group (for instance, inotropes or vasopressors) and noting pre-ECMO temporary mechanical support. Pre-ECMO infectious organisms were grouped into bacterial, fungal and viral. A combined 105 continuous, categorical, ordinal, and binary variables were included in the analysis (Supplementary Materials 2—eTable1).
Continuous data were checked for outliers and probable erroneous entries by hard data limits as agreed by clinical consensus (MS, AD—Supplementary Materials 2—eTable 2). Data outside these limits were truncated or removed for later imputation. Missingness of data varied widely across all variables, which is typical for large clinical cohorts [26, 27] (Supplementary Materials 2—eTable 1). Variables with more than 50% missingness were removed entirely based on preliminary experiments exploring feature importance against model performance [28]. Missingness was attributed to missing at random or not at random; for example, patients with extracorporeal cardiopulmonary resuscitation (ECPR) were more likely to have missingness on pre-ECMO biochemistry values. All data were min–max scaled, and missing data were imputed using a Gaussian Copula method, chosen due to its proficiency at handling highly mixed variables (including categorical, ordinal, and continuous variables) and non-normally distributed data [29, 30]. Outcomes data were only modestly unbalanced, with 41.3% survival to hospital discharge, in keeping with global adult VA-ECMO survival rates [31]; as such, resampling was not performed.
Machine learning methods
Data were applied to seven AI and ML methods: Decision Tree, Logistic Regression, Random Forest Ensemble, AdaBoost Ensemble, Extreme Gradient Boosted Ensemble, Support Vector Machine, and Neural Network to find the best classifier for predicting in-hospital mortality (primary endpoint). After extensive hyperparameter tuning and feature selection, a Deep Neural Network (DNN) was found to best predict in-hospital mortality (additional details in Supplementary Materials 1). Briefly, a DNN mimics the behaviour of human learning, recognising patterns to identify and solve problems. It does so using input features (patient variables) which are passed to neurons; if the feature is prominent enough, the neuron is “activated”, and the output is passed to the next layer, where the feature is again analysed in combination with the other features passed forward. Once all layers have been activated, the DNN decides the outcome, in this case, survival or in-hospital mortality [32]. The DNN then reinforces neurons that make correct predictions, strengthening the behaviour. Neural Networks are considered “deep” if there is more than one activation layer [33].
Input features that were predictive for the DNN, named ECMO Predictive Algorithm (ECMO PAL), were selected through permutation feature importance from the six remaining models, keeping variables with a predictive power > 0.001, and comparing Pearson correlation coefficients to determine co-linearity between those features. Feature selection was done using the other ML models due to the long (hours) training time required for feature selection using the DNN directly. Hyperparameter optimisation revealed that a four-layer DNN with 20% dropout (which randomly censors 20% of the data during training) provided the best outcomes across fivefold cross-validation. The trained DNN was evaluated internally and externally and compared against other published VA-ECMO outcomes scores.
Model evaluation
Shapley Additive Explanations (SHAP) were used to understand feature importance and illustrate each feature’s effect on model predictions [34], while partial dependence plots were used to explore variable interactions in more detail [35,36,37].
Internal model validation was achieved using fivefold cross-validation. In-hospital mortality cutoff was defined as a survival prediction of less than 50%. The key values of interest were sensitivity (ratio of correct mortality predictions to observed mortality: true positives/true positives + false negatives) and precision (positive predictive value − ratio of correct mortality predictions to total mortality predictions: true positives/true positives + false positives) to in-hospital mortality and overall model accuracy (ratio of correct predictions to incorrect predictions) and area under the receiver operating characteristic (AUROC) to allow direct comparison with other published models.
External validation was performed on a new epoch of data from the ELSO database (all adult VA-ECMO patients in 2021). This epoch incorporated all pertinent regions and quantified model drift with time. The external validation data initially consisted of 6210 patients. The training data exclusion criteria were applied to the validation data resulting in 5015 patients from 387 centres for analysis. The validation data were cleaned, imputed, and scaled in the same manner as the training data and predictions were made by ECMO PAL, assessing the same metrics as with training. Finally, ECMO PAL predictions were compared against previously published VA-ECMO survival scores: SAVE Score [20], Modified SAVE Score [9], and ECMO-ACCEPTS [7] chosen based on available variables within the ELSO registry. The comparator scores made predictions on the external validation data based on the scope of the score (cardiogenic shock patients for SAVE and Modified SAVE; all patients for ECMO-ACCEPTS). The AUROCs and overall accuracy of each score were then directly compared. Where appropriate, data are presented as mean ± standard deviation.
Results
Training patient demographics and clinical data
Following exclusion, 18,167 patients were included for model training (Fig. 1). The patient mean age was 56.8 ± 15.1 years, with 66.7% male and 33.3% female (Table 1). Patient races were recorded as Asian (14.7%), Black (10%), Hispanic (5%), Multiple (6.2%), White (55.5%), Other (3.8%), and Unknown (4.8%). Patients came from 467 ECMO centres across the established ELSO regions of Asia–Pacific, Europe, Latin America, North America, and South and West Asia and Africa. No socioeconomic data were available. In addition, 50.2% of patients had a cardiac arrest within 24 h before ECMO initiation, while 26% had ECPR as their primary ECMO mode. The distribution of comorbidities is listed in detail in the online materials (Supplementary Materials 2—eTable 1).
Feature importance and explainers
Permutation feature importance and Pearson correlation were used to identify and reduce the model input variables. Systolic and diastolic blood pressure were highly correlated, as were partial pressure of arterial oxygen (PaO2) and arterial oxygen saturation (SaO2), pH and lactate. No loss in accuracy occurred when excluding diastolic blood pressure and SaO2, which were removed. However, removing pH or lactate resulted in losses, and as such, both were retained in the model. Through this process, 105 features were reduced to 23 features with predictive power and the DNN feature importance was defined post-hoc using SHAP explainers (Fig. 2).
Shapley Additive Explainer (SHAP) values show the influence of each variable on the model output. The colour of each dot represents the value of an individual patient data point, with pink being the maximum variable value and blue being the minimum variable value. The dot’s position on the x-axis represents that data point’s contribution to a patient-specific outcome in combination with all other variables for that patient. The width of the violin shows the distribution. Variables are ranked in descending order of predictive importance
The most predictive variables were lactate, age, serum bicarbonate (HCO3), respiratory rate, and pre-ECMO endotracheal intubation time. Many variables had a clear monotonic relationship to the outcome. For example, higher age, lactate, and time intubated before ECMO were all monotonically associated with lower survival. Conversely, higher PaO2 and administration of pre-ECMO bicarbonate and inotropic infusions were all monotonically associated with higher survival. This can be seen by the gradual change from pink to blue of such variables in Fig. 2. Several variables had non-monotonic effects on patient outcomes, for example, HCO3, pH, and systolic blood pressure, reflecting an ideal range for those variables, as demonstrated by the pink and blue dots scattered across the x-axis in Fig. 2. A lower respiratory rate was found to be more predictive of patient mortality, and patients with a respiratory rate of less than five breaths/second had the lowest mortality, likely representing un-intubated patients in cardiac arrest at ECMO initiation. Conversely, higher respiratory rates had reduced mortality, possibly representing patients with lower sedation and spontaneously breathing. Dichotomous variables such as heart failure and cardiac arrest both increased and decreased mortality predictions depending on the patient, indicating a complex interaction with other patient variables and highlighting possible patient selection biases inherent with eligibility decisions around ECMO (Fig. 2). Interactions between variables were further investigated using partial dependence plots; for example, a complex interaction was found between cardiac arrest status and lactate (Fig. 3). Additional information about the variable impact on model predictions can be found in Supplementary Materials 1.
Partial dependence plot showing patient cardiac arrest status and interaction with lactate. Yellow contour regions have the highest survival, while dark purple regions have the lowest survival rates. Large black ticks at the bottom represent the deciles of the data. Contour labels represent the effect on survival. ECPR extracorporeal membrane oxygenation cardiopulmonary resuscitation
Internal and external validation
Fivefold internal validation of the model was conducted, demonstrating strong sensitivity and precision to in-hospital mortality, 82.1 ± 0.2% and 77.6 ± 0.2%, respectively (Table 2). Overall model accuracy was 75.5 ± 0.1%. External validation was conducted on 5015 patients, with in-hospital mortality sensitivity and precision marginally dropping by 4.4% and 4.2%, respectively, to 77.7% and 73.4% (Table 2). Overall accuracy reduced to 72.7% [99% CI 71.1–74.3], exhibiting only a 2.8% drop in accuracy between internal and external validation. The model demonstrated consistent performance regardless of VA-ECMO indication (accuracy within 3%), with the highest sensitivity to in-hospital mortality for an indication of toxins (85.7%) and the lowest sensitivity for patients with myocarditis (73.2%, Table 3). Ground truth in-hospital mortality was 58.7% in the training cohort and 54.4% in the validation cohort. The model calibration curve yielded a Brier Score of 0.22 (Supplementary Materials 1).
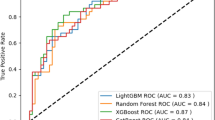
The SAVE score, Modified SAVE score, and ECMO ACCEPTS scores were compared against ECMO PAL. Predictions were made for all 5,015 validation patients for ECMO ACCEPTS, and 4476 patients with cardiogenic shock for SAVE and modified SAVE (cardiogenic shock derived scores). ECMO ACCEPTS demonstrated the lowest performance on the validation data (Accuracy: 54.7%, AUROC: 0.61), followed by the SAVE score (Accuracy: 61.1%, AUROC: 0.61), modified SAVE SCORE (Accuracy: 62%, AUROC: 0.67), and ECMO PAL (Accuracy: 72.7%, AUROC: 0.80) (Supplementary Fig. 1). For direct reference, ECMO PAL also made predictions on the 4476 cardiogenic shock patients only, and the accuracy and AUROC were within the stated confidence intervals (Table 2) (Supplementary Material 2—eTable 3).
Discussion
This study investigated a novel AI-driven tool for predicting in-hospital mortality of patients receiving VA-ECMO. Leveraging big data from the international ELSO registry, ECMO PAL was derived on 18,167 patients (2017–2020) and validated on 5015 additional patients (2021). ECMO PAL outperformed existing scores in both accuracy and AUROC when predicting outcomes of a large international cohort. ECMO PAL was most sensitive and precise at predicting in-hospital mortality.
ECMO PAL was compared with established ECMO outcomes scores and found to have superior accuracy and AUROC. Only one other AI-powered ECMO model has been published in the past. Ayers et al. trained a DNN to predict survival outcomes, reporting an accuracy of 82% and an AUROC of 0.9 [5]. That study did not include patients who survived for less than 48 h. That model was also trained on 197 patients and evaluated on 50 patients from a single centre, likely resulting in low generalisability. Direct comparison of outcomes scores must be interpreted cautiously as many scores are derived for specific regions, a specific disease aetiology, or specific ECMO indication. Those specific models will perform better in their researched context but may not be generalisable to other health regions or patient cohorts. In contrast, ECMO PAL is highly generalisable, having been trained on patients from 467 ECMO centres across Europe, Asia–Pacific, North America, Latin America, and Southwest Asia and Africa. This high generalisability highlights the model’s potential for risk adjustment, quality control, and embedding into national and international registries.
As clinical practice and patient populations evolve, the performance of clinical prediction models may deteriorate over time, and a 2.8% drop in performance was seen in ECMO PAL between 2017–2020 and 2021. In the case of ECMO PAL, an infrastructure has been established to retrain and update the model with new data and predictive variables as they become available, with major updates planned for every 2–3 years. Meanwhile, tracking the year within the model allows for annual calibration adjustment reflecting trends in reduced mortality outcome (0.2–0.5% each year) between major model updates. In this regard, ECMO PAL is an evolving algorithm that can continue to learn, allowing for ongoing optimisation of its prediction performance and clinical utility.
SHAP explainers were used to understand the effects of different variables on the predicted outcome. Many variables were found to be monotonically influential on the predicted outcome and have been reported in previous models, such as age, lactate, and pre-ECMO intubation time [9, 12, 20]. The effect of some variables is clinically intuitive, with older age and longer intubation times associated with worse outcomes. However, this model also highlighted complex interactions with other variables which were not monotonically associated with outcome and are less straightforward to interpret clinically. For instance, pre-ECMO cardiac arrest and ECPR contributed to both survival and non-survival predictions in different individual patients. Further investigation using partial dependence plots revealed that those with low pre-ECMO lactate had the highest survival regardless of arrest status (Fig. 3), likely representing a set of patients who were initiated on ECMO very quickly after cardiac arrest. Meanwhile, patients with cardiac arrest or ECPR who had elevated lactates (8–30 mmol/L) performed poorly. Interestingly, no patients with extremely high lactates (35–40 mmol/L) were recorded as having a cardiac arrest, potentially indicating some other underlying pathology, data entry error, or patient selection bias. Although included in the initial analysis, features such as gender, VA-ECMO indication, and comorbidities were not considered highly predictive of outcome. Low model importance does not mean these features are not clinically meaningful or have no bearing on the outcome; rather, it illustrates that these features are likely already encapsulated by the predictive demographic, clinical, and biochemistry data.
Specific indications for ECMO initiation were included in the analysis but were found to have a limited impact on model prediction. Except for pulmonary embolism, including ECMO indication did not improve model performance. Instead, it is likely that the model has already captured the determinants associated with disease/indication-specific outcomes. In this regard, the benefit of AI-powered tools is highlighted, in which there is capacity to look beyond human-defined classifications and into the complex interplay of variables found in the numerical data (clinical, biochemistry, haemodynamic). Of note, while not reported in detail here, an ensemble modelling method was investigated during this research, whereby training individual models for each identified ECMO indication was performed. These disease-specific models had lower individual accuracies than the generalised model, likely due to the lower number of patients available for training each model.
A limitation of this model was the selection bias inherent in the training and validation data. The ELSO registry records only patients supported with ECMO and does not include critically ill patients for whom clinicians elected not to initiate ECMO. Thus, limiting the utility of ECMO PAL for decision-making related to ECMO initiation. Nevertheless, globally, different hospitals will likely have different thresholds for commencing ECMO. Thus, this variation in the threshold may be reflected in the model using a large international cohort. Although ECMO-PAL may not yet be mature enough to make individual-level predictions for resource allocation, the model could be used for cohort predictions in risk adjustment and quality control.
A further limitation of the model is the varying quality of available data, with different centres having a higher and lower propensity to enter erroneous or irrelevant data. For instance, some centres assigned over 90 comorbidities for a single patient as contributing to their reason for being on ECMO, adding additional noise to the data set. Conversely, only 40% of the cohort had one or more secondary diagnoses or comorbidities recorded. There were also modest (up to 44%) levels of missing data for some key variables, such as respiratory rate and blood gas parameters, including lactate. These data were deemed missing not at random due to certain disease aetiologies, where these data would not be routinely recorded (ECPR, for example). Robust imputation methods were used to fill the data; however, imputation is imperfect, and this may affect model real-world model performance. More, higher quality data will remove the need for imputation in future model updates.
In addition, assumptions were made during data cleaning. For example, data from International Classification of Diseases tenth revision (ICD-10) codes, procedure codes, and pre-ECMO support were operationalised into categorical and binary variables for supervised learning. The process of operationalising this data was done manually and was based on the experience of multiple clinicians, with room for interpretation. A common narrative is that the black-box nature of neural networks will limit their clinical acceptability [38]. This limitation may be overstated [39], as although the interdependence of the model variables and their effect on outcomes is somewhat opaque to the user, explainers such as SHAP and partial dependence plots can help understand the model outputs at both a general and patient-specific level.
Clinical outcome data post-discharge was unavailable via the ELSO registry, and as a result, this study did not investigate predictors of longer term patient outcomes.
ECMO PAL is the first step in a suite of evolving AI-powered tools for prediction in ECMO. ECMO PAL will be implemented as a free online web app (ecmo-pal.icu). Notably, the web app uses individual patient explainers to understand the prediction made for the inputted patient (Supplementary Fig. 2), allowing a deeper understanding of each outcome prediction. More broadly, this work demonstrates the potential of AI for ICU outcomes prediction. The accuracy of AI-powered tools for ECMO can be further improved by modifying registry data collection processes to optimise its use in big data analysis. A key example would be transitioning from ICD-10 codes for patient diagnoses and creating a list of acceptable primary diagnoses and common comorbidities instead.
Conclusion
This study aimed to create a novel AI-driven tool for predicting in-hospital mortality in patients receiving venoarterial ECMO. The developed model demonstrated high generalisability across regions and made accurate predictions across a heterogeneous patient cohort. This study is the first to investigate AI-driven tools for ECMO using global big data. This research forms the starting point for ECMO PAL, specifically designed to be updated and evolve as ECMO therapy matures. As medicine advances through the digital age, AI-powered clinical prediction tools such as ECMO PAL present a platform to enhance patient prognostication, improve resource allocation, and identify solutions for optimising patient care.
Data availability
This article’s data cannot be shared publicly as it is private health data contributed by participating hospitals to the Extracorporeal Life Support Organization. Data requests should be addressed directly to ELSO (elso.org).
References
Combes A, Price S, Slutsky AS, Brodie D (2020) Temporary circulatory support for cardiogenic shock. Lancet 396:199–212. https://doi.org/10.1016/S0140-6736(20)31047-3
Harvey MJ, Gaies MG, Prosser LA (2015) US and international in-hospital costs of extracorporeal membrane oxygenation: a systematic review. Appl Health Econ Health Policy 13:341–357. https://doi.org/10.1007/s40258-015-0170-9
Rozencwajg S, Fraser J, Montero S et al (2017) To be or not to be on ECMO: can survival prediction models solve the question? Crit Care Resusc 19:21–28
Wengenmayer T, Duerschmied D, Graf E et al (2019) Development and validation of a prognostic model for survival in patients treated with venoarterial extracorporeal membrane oxygenation: the PREDICT VA-ECMO score. Eur Hear J Acute Cardiovasc Care 8:350–359. https://doi.org/10.1177/2048872618789052
Ayers B, Wood K, Gosev I, Prasad S (2020) Predicting survival after extracorporeal membrane oxygenation using machine learning. Ann Thorac Surg. https://doi.org/10.1016/j.athoracsur.2020.03.128
Mégarbane B, Deye N, Aout M et al (2011) Usefulness of routine laboratory parameters in the decision to treat refractory cardiac arrest with extracorporeal life support. Resuscitation 82:1154–1161. https://doi.org/10.1016/j.resuscitation.2011.05.007
Becher PM, Twerenbold R, Schrage B et al (2020) Risk prediction of in-hospital mortality in patients with venoarterial extracorporeal membrane oxygenation for cardiopulmonary support: the ECMO-ACCEPTS score. J Crit Care 56:100–105. https://doi.org/10.1016/j.jcrc.2019.12.013
Peigh G, Cavarocchi N, Keith SW, Hirose H (2015) Simple new risk score model for adult cardiac extracorporeal membrane oxygenation: simple cardiac ECMO score. J Surg Res 198:273–279. https://doi.org/10.1016/j.jss.2015.04.044
Chen WC, Huang KY, Yao CW et al (2016) The modified SAVE score: predicting survival using urgent veno-arterial extracorporeal membrane oxygenation within 24 hours of arrival at the emergency department. Crit Care 20:1–7. https://doi.org/10.1186/s13054-016-1520-1
Schmidt M, Bailey M, Sheldrake J et al (2014) Predicting survival after extracorporeal membrane oxygenation for severe acute respiratory failure: the respiratory extracorporeal membrane oxygenation survival prediction (RESP) score. Am J Respir Crit Care Med 189:1374–1382. https://doi.org/10.1164/rccm.201311-2023OC
Muller G, Flecher E, Lebreton G et al (2016) The ENCOURAGE mortality risk score and analysis of long-term outcomes after VA-ECMO for acute myocardial infarction with cardiogenic shock. Intensive Care Med 42:370–378. https://doi.org/10.1007/s00134-016-4223-9
Wang L, Yang F, Wang X et al (2019) Predicting mortality in patients undergoing VA-ECMO after coronary artery bypass grafting: the REMEMBER score. Crit Care 23:1–10. https://doi.org/10.1186/s13054-019-2307-y
Maslove DM, Elbers PWG, Clermont G (2021) Artificial intelligence in telemetry: what clinicians should know. Intensive Care Med 47:150–153. https://doi.org/10.1007/s00134-020-06295-w
Mamdani M, Slutsky AS (2021) Artificial intelligence in intensive care medicine. Intensive Care Med 47:147–149. https://doi.org/10.1007/s00134-020-06203-2
Bossuyt PM, Reitsma JB, Bruns DE et al (2015) STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem 61:1446–1452
Hernandez-Boussard T, Bozkurt S, Ioannidis JPA, Shah NH (2020) MINIMAR (MINimum information for medical AI reporting): developing reporting standards for artificial intelligence in health care. J Am Med Inform Assoc 27:2011–2015
Collins GS, Reitsma JB, Altman DG, Moons KGM (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) the TRIPOD statement. Circulation 131:211–219. https://doi.org/10.1161/CIRCULATIONAHA.114.014508
The Extracorporeal Life Support Organization ELSO Registry. https://www.elso.org/Registry.aspx
Manchikanti L, Falco FJE, Hirsch JA (2013) Ready or not! Here comes ICD-10. J Neurointerv Surg 5:86–91. https://doi.org/10.1136/neurintsurg-2011-010155
Schmidt M, Burrell A, Roberts L et al (2015) Predicting survival after ECMO for refractory cardiogenic shock: the survival after veno-arterial-ECMO (SAVE)-score. Eur Heart J 36:2246–2256. https://doi.org/10.1093/eurheartj/ehv194
Thiagarajan RR, Barbaro RP, Rycus PT et al (2017) Extracorporeal life support organization registry international report 2016. ASAIO J 63:60–67. https://doi.org/10.1097/MAT.0000000000000475
Eckman PM, Katz JN, El Banayosy A et al (2019) Veno-arterial extracorporeal membrane oxygenation for cardiogenic shock: an introduction for the busy clinician. Circulation 140:2019–2037. https://doi.org/10.1161/CIRCULATIONAHA.119.034512
Charlson ME, Pompei P, Ales KL, MacKenzie CR (1987) A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. https://doi.org/10.1016/0021-9681(87)90171-8
Toson B, Harvey LA, Close JCT (2016) New ICD-10 version of the multipurpose australian comorbidity scoring system outperformed Charlson and Elixhauser comorbidities in an older population. J Clin Epidemiol 79:62–69. https://doi.org/10.1016/j.jclinepi.2016.04.004
Thorwarth WT (2004) From concept to CPT code to compensation: how the payment system works. J Am Coll Radiol 1:48–53. https://doi.org/10.1016/S1546-1440(03)00020-6
El-Rashidy N, El-Sappagh S, Abuhmed T et al (2020) Intensive care unit mortality prediction: an improved patient-specific stacking ensemble model. IEEE Access 8:133541–133564. https://doi.org/10.1109/ACCESS.2020.3010556
Jeffery AD, Dietrich MS, Fabbri D et al (2018) Advancing in-hospital clinical deterioration prediction models. Am J Crit Care 27:381–391. https://doi.org/10.4037/ajcc2018957
Vesin A, Azoulay E, Ruckly S et al (2013) Reporting and handling missing values in clinical studies in intensive care units. Intensive Care Med 39:1396–1404. https://doi.org/10.1007/s00134-013-2949-1
Zhao Y, Udell M (2020) Missing value imputation for mixed data via Gaussian copula. In: Proceedings of ACM SIGKDD international conference on knowledge discovery and data mining, pp 636–646. https://doi.org/10.1145/3394486.3403106
Zhao Y, Landgrebe E, Shekhtman E, Udell M (2022) Online missing value imputation and change point detection with the Gaussian copula. Proc AAAI Conf Artif Intell 36:9199–9207. https://doi.org/10.1609/aaai.v36i8.20906
Extracorporeal Life Support Organization (2020) International Registry Summary (July - 2020). elso.org/registry/internationalsummaryandreports/internationalsummary.aspx. Accessed 19 March 2021
Georgevici AI, Terblanche M (2019) Neural networks and deep learning: a brief introduction. Intensive Care Med 45:712–714. https://doi.org/10.1007/s00134-019-05537-w
Brownlee J (2016) Machine learning algorithms from scratch with Python. Machine Learning Mastery. p 237
Lundberg SM, Lee SI (2017) A unified approach to interpreting model predictions. Adv Neural Inf Process Syst 2017:4766–4775
Zhao X, Yang H, Yao Y et al (2022) Factors affecting traffic risks on bridge sections of freeways based on partial dependence plots. Phys A Stat Mech Appl 598:127343. https://doi.org/10.1016/j.physa.2022.127343
Scikit-learn Developers (2023) Partial dependence plots and individual conditional expectation plots. https://scikit-learn.org/stable/modules/partial_dependence.html. Accessed 21 June 2023
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12(85):2825–2830
Wang F, Kaushal R, Khullar D (2020) Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann Intern Med 172:59. https://doi.org/10.7326/M19-2548
Durán JM, Jongsma KR (2021) Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J Med Ethics 47:329–335. https://doi.org/10.1136/medethics-2020-106820
Acknowledgements
The authors would like to acknowledge the significant contributions from the participating Extracorporeal Life Support Organisation centres. The administrative support from the Extracorporeal Life Support Organisation (ELSO) and Australian & New Zealand Intensive Care Research Centre (ANZIC-RC), as well as the feedback given by the International ECMO Network (ECMONet) Scientific Committee. The authors acknowledge Ming Sun for his contributions to the early planning of this research.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by Monash University, the Baker Heart and Diabetes Institute, and the Extracorporeal Life Support Organisation. CH and AS were supported by the National Health and Medical Research Council (CRE ICU). SG was supported by the National Heart Foundation of Australia (#106675) and the National Health and Medical Research Council (#2016995).
Author information
Authors and Affiliations
Consortia
Contributions
All authors were involved in the planning and execution of the research. AS and MS did the machine learning. MS, DP, AD, and RB provided detailed reviews for clinical consensus. CH, DB, DK, and DP provided statistical feedback. CH, RB, DP, and VP provided registry support. SG provided engineering support. AS and MS wrote the first draft and all other authors provided critical review.
Corresponding author
Ethics declarations
Conflicts of interest
DB received research support from and consults for LivaNova. He has been on the medical advisory boards for Abiomed, Xenios, Medtronic, Inspira and Cellenkos. He is the President-elect of the Extracorporeal Life Support Organization (ELSO) and the Chair of the Executive Committee of the International ECMO Network (ECMONet). RB is a member of the Extracorporeal Life Support Organisation steering committee. In addition, CH, AS, VP, and AD, are ELSO members. CH and VP are members of the international ECMO Network Executive Committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Stephens, A.F., Šeman, M., Diehl, A. et al. ECMO PAL: using deep neural networks for survival prediction in venoarterial extracorporeal membrane oxygenation. Intensive Care Med 49, 1090–1099 (2023). https://doi.org/10.1007/s00134-023-07157-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-023-07157-x