Abstract
Aims/hypothesis
Regulatory factor X 6 (RFX6) is crucial for pancreatic endocrine development and differentiation. The RFX6 variant p.His293LeufsTer7 is significantly enriched in the Finnish population, with almost 1:250 individuals as a carrier. Importantly, the FinnGen study indicates a high predisposition for heterozygous carriers to develop type 2 and gestational diabetes. However, the precise mechanism of this predisposition remains unknown.
Methods
To understand the role of this variant in beta cell development and function, we used CRISPR technology to generate allelic series of pluripotent stem cells. We created two isogenic stem cell models: a human embryonic stem cell model; and a patient-derived stem cell model. Both were differentiated into pancreatic islet lineages (stem-cell-derived islets, SC-islets), followed by implantation in immunocompromised NOD-SCID-Gamma mice.
Results
Stem cell models of the homozygous variant RFX6−/− predictably failed to generate insulin-secreting pancreatic beta cells, mirroring the phenotype observed in Mitchell–Riley syndrome. Notably, at the pancreatic endocrine stage, there was an upregulation of precursor markers NEUROG3 and SOX9, accompanied by increased apoptosis. Intriguingly, heterozygous RFX6+/− SC-islets exhibited RFX6 haploinsufficiency (54.2% reduction in protein expression), associated with reduced beta cell maturation markers, altered calcium signalling and impaired insulin secretion (62% and 54% reduction in basal and high glucose conditions, respectively). However, RFX6 haploinsufficiency did not have an impact on beta cell number or insulin content. The reduced insulin secretion persisted after in vivo implantation in mice, aligning with the increased risk of variant carriers to develop diabetes.
Conclusions/interpretation
Our allelic series isogenic SC-islet models represent a powerful tool to elucidate specific aetiologies of diabetes in humans, enabling the sensitive detection of aberrations in both beta cell development and function. We highlight the critical role of RFX6 in augmenting and maintaining the pancreatic progenitor pool, with an endocrine roadblock and increased cell death upon its loss. We demonstrate that RFX6 haploinsufficiency does not affect beta cell number or insulin content but does impair function, predisposing heterozygous carriers of loss-of-function variants to diabetes.
Data availability
Ultra-deep bulk RNA-seq data for pancreatic differentiation stages 3, 5 and 7 of H1 RFX6 genotypes are deposited in the Gene Expression Omnibus database with accession code GSE234289. Original western blot images are deposited at Mendeley (https://data.mendeley.com/datasets/g75drr3mgw/2).
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
Diabetes is a metabolic disorder characterised by the inability of pancreatic beta cells to control blood glucose levels. The aetiology of the most common form, type 2 diabetes, is heterogeneous, although population genetic studies have identified numerous type 2 diabetes-associated genetic variants in loci of genes expressed in the beta cell [1, 2]. Pathogenic variants in these beta cell genes lead to monogenic diabetes, presenting either as neonatal diabetes or MODY [3]. Regulatory factor X 6 (RFX6) is a winged-helix transcription factor that regulates genes required for the development of the pancreas and other intestinal organs [4,5,6,7]. Autosomal recessive variants in RFX6 cause Mitchell–Riley syndrome (OMIM, 615710; www.omim.org), characterised by intrauterine growth retardation, annular or hypoplastic pancreas, permanent neonatal diabetes, gall bladder hypoplasia or agenesis, intestinal stenosis with malabsorptive diarrhoea and, in some cases, pancreatic exocrine insufficiency [6, 8, 9]. Recently, a less severe manifestation of Mitchell–Riley syndrome was documented, involving compound heterozygous variants that are not fully inactivating. This condition presents as childhood-onset diabetes, typically occurring between the ages of 2 and 5 years [10, 11].
In mice, Rfx6 is initially expressed in the definitive endoderm, before becoming progressively confined to the gut and dorsal pancreatic bud, and then islet progenitor cells where it is required for the differentiation of all islet cell types, except for pancreatic polypeptide (PP)-producing cells. Mice with loss of Rfx6 exhibited similarities to the human phenotype, presenting with neonatal diabetes and intestinal obstruction, albeit with variable pancreatic hypoplasia [6]. While homozygous Rfx6 mutant mice had severe symptoms and died shortly after birth, heterozygous mutants did not show signs of diabetes [6]. Conditional Rfx6 ablation in adult mouse beta cells resulted in impaired glucose-stimulated insulin secretion (GSIS), without compromised beta cell mass or insulin content. The defective insulin secretion was attributed to the downregulation of beta cell maturation genes Gck, Abcc8, Ucn3 and voltage-dependent calcium channel (VDCC) genes, concomitant with the upregulation of beta cell disallowed genes such as Slc16a1, Ldha, and Igfbp4 [12]. Similarly, knockdown of RFX6 in the human beta cell insulinoma line EndoC-βH2 resulted in reduced insulin gene transcription and defective GSIS through reducing VDCC gene expression [13]. A recent study also demonstrated that RFX6 knockdown in primary human islets reduced GSIS to the level seen in islets from donors with type 2 diabetes, through transcriptionally dysregulated vesicle trafficking, exocytosis and ion transport pathways [14].
Heterozygous RFX6 pathogenic variants have been linked to MODY with reduced penetrance in humans [15,16,17,18,19,20,21]. Moreover, genome-wide association studies have associated variants of RFX6 with type 2 diabetes [22, 23]. In adult primary islets, RFX6 was shown to be a hub transcription factor that was downregulated in islets from donors with early type 2 diabetes, correlated with reduced GSIS [14]. Additionally, genetic variants that increase type 2 diabetes risk are predicted to disrupt RFX-binding motifs [24].
The precise mechanism of how heterozygous pathogenic RFX6 variant carriers are predisposed to develop diabetes remains unknown. Therefore, we sought to use patient-derived and embryonic stem cells combined with CRISPR-based genetic engineering to create isogenic allelic series models of a specific RFX6 frameshift variant, circumventing the use of unphysiological systems of complete gene knockout or knockdown. Human pluripotent stem cells have been extensively used to model monogenic diabetes genes [25], including RFX6 [9, 26, 27], as they can be differentiated into stem-cell-derived islets (SC-islets) that closely mimic native human islets developmentally and functionally. Employing our optimised protocol to generate highly functional SC-islets [28, 29], we elucidate the impact of homozygous and heterozygous RFX6 pathogenic variants on pancreatic endocrine development and beta cell function.
Methods
Lifetime risk analysis of type 2 and gestational diabetes in the FinnGen dataset
We analysed the risk of gestational or type 2 diabetes, the age at diagnosis of diabetes and BMI in the RFX6 frameshift variant (p.His293LeufsTer7) carriers compared with non-carriers in the FinnGen data freeze R11 at individual level, using genome-wide genotype data and longitudinal healthcare registry data [23, 30]. FinnGen (https://www.finngen.fi/en) is a public–private research project, combining genome and digital healthcare data gathered since 2017. In FinnGen, diseases are defined as endpoints by FinnGen clinical expert teams according to ICD codes of versions 8 (1969–1986), 9 (1987–1995; http://www.icd9data.com/2007/Volume1/default.htm) and 10 (1996–2019; https://icd.who.int/browse10/2019/en). FinnGen R11 comprises 440,734 Finnish European individuals (44% male and 56% female) with informed consent for biobank research based on the Finnish Biobank Act, or regarding earlier cohorts, with approval from Fimea, the National Supervisory Authority for Welfare and Health. As the database is based in Finland, where the variant under study is enriched, the majority of samples belong to Finnish ethnicity, with no exclusions made based on gender or sex. The type 2 diabetes population comprised 55% male and 45% female individuals. The FinnGen study protocol (no. HUS/990/2017) is approved by the Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS).
Generating induced pluripotent stem cells from an individual with Mitchell–Riley syndrome, and stem cell culturing
Dermal fibroblasts obtained from a skin biopsy from an individual with homozygous RFX6 frameshift variant at 6 months of age were reprogrammed using Sendai viruses (SeVdp), expressing OCT4, SOX2, KLF4 and C-MYC reprogramming factors. The formed HEL118 induced pluripotent stem cell (iPSC) lines were verified using Sanger sequencing to confirm the frameshift variant, were tested negative for mycoplasma contamination and did not contain transgene vectors. Additionally, the iPSCs line HEL46.11 (derived from human neonatal foreskin fibroblast) and the human embryonic stem cell (hESC) line H1 (WA01, WiCell; Madison, USA), which are wild-type RFX6, were used in this study. All the generated stem cell lines were cultured on Matrigel-coated (Corning, no. 354277; USA) plates with E8 medium (Thermo Fisher Scientific, A1517001; USA) and passaged using 0.5 mmol/l EDTA (Life Technologies, 15575–038; USA) in PBS as a dissociation agent. The patient’s iPSCs were derived after informed consent from parents and according to the approval of the coordinating ethics committee of the Helsinki and Uusimaa Hospital District (no. 423/13/03/00/08). The patient’s clinical data were used after informed consent from parents.
Genome editing of iPSCs and H1 hESCs
For correcting the RFX6 frameshift variant in the patient-derived HEL118.3 iPSCs, we used a plasmid-based CRISPR–Cas9 system with a guide and a correction template that were designed with Benchling (Biology Software, 2017, https://benchling.com) (gRNA1 correction: ATAACAGGATTTTCGAGCAG). The correction template introduced a TseI restriction site by silent mutations to facilitate the screening of the edited clones. Two million iPSCs were electroporated with 6 µg of CAG-Cas9-T2A-EGFP-ires-Puro (Addgene, plasmid no. 78311), 500 ng of the gRNA cassette and 4 µg of the 200 bp dsDNA correction template, using Neon Transfection system (Thermo Fisher Scientific; 1100 V; 20 ms; two pulses). Single cells were sorted, expanded and screened using TseI restriction enzyme. The editing resulted in heterozygous clones only; one heterozygous clone was electroporated using gRNA2 correction guide (AGCAGGGGAAGGAGATGGTC) to obtain the homozygous clone. All clones (iPSCs; #RFX6−/−, #RFX6+/− and #RFX6+/+) were further validated by Sanger sequencing.
For introducing the RFX6 frameshift variant in H1 hESCs, we used a more efficient ribonucleoprotein (RNP) CRISPR–Cpf1 system (Integrated DNA Technologies, USA), with a guide and a mutation template that were designed with Benchling (Biology Software, 2017) (gRNA mutation: TTACACTTTTGGCAAGGAATG). Two million cells were electroporated with 10 µg Alt-R A.s. Cas12a (Cpf1) Ultra combined with the gRNA and 4 µg of the 100-nt ssODN correction template (Integrated DNA Technologies), using Neon Transfection system (Thermo Fisher Scientific; 1100 V; 20 ms; two pulses). Single cells were sorted, expanded and screened using TseI restriction enzyme. Since the cut-site is 13 nucleotides away from the mutation, the suboptimal editing resulted in heterozygous clones only. The heterozygous clone 3G was electroporated with the same gRNA and a mutation template carrying TseI and NheI restriction sites to facilitate the screening of homozygous clones. All clones (hESCs; RFX6+/+, RFX6+/− 9C, RFX6+/− 3G and RFX6−/−) were further validated by Sanger sequencing. The top three CRISPR off-target hits were sequenced and did not have any indels. Karyotype analyses based on chromosomal G-banding were performed for all the generated hiPSC and hESC clones at Ambar Lab, Barcelona, Spain. All cell lines tested negative for mycoplasma. Primer sequences used for genome editing are described in electronic supplementary material (ESM) Table 1.
In-vitro pancreatic endocrine differentiation culture
HEL118.3 iPSCs were differentiated towards the pancreatic lineage to generate pancreatic endocrine cells using our previously published protocol with minor modifications [31]. Briefly, two million cells/well were seeded on six-well Matrigel-coated plates in E8 medium supplemented with 10 µmol/l Rho-Associated kinase inhibitor (ROCKi, Y-27632). The differentiation was started following 24 h of seeding and proceeded through a six-stage differentiation protocol in monolayer. The hESC H1 cell lines were differentiated using the extended seven-stage protocol (stages 1–7 [S1–S7]: S1–S4 in adherent culture; S5 in AggreWell [Stemcell Technologies, no. 34421; Vancouver, Canada] and S6 and S7 in suspension culture) [28, 29]. The detailed differentiation protocol and the stage-specific complete media formulations are described in ESM Tables 2, 3. The details of the reagents used in the differentiation protocol are described in ESM Table 4.
Western blot
For protein extraction, cells were washed with ice-cold PBS and lysed with Cell lysis buffer (Cell Signalling Technology no. 9803; USA) for 10 min on ice. The cells were sonicated for 3×5 s on ice, centrifuged (10,000 g for 10 min at 4°C) and the supernatant fraction was stored at −80°C. The samples were run on Any kD Mini-PROTEAN TGX gel (Bio-Rad, Hercules, USA) and then dry-transferred onto a nitrocellulose membrane using the iBlot system (Invitrogen; USA) as per manufacturer’s instructions. The membrane was then probed with the primary antibody overnight at 4°C, washed twice with TBS (Medicago no. 09-7500; Uppsala, Sweden) containing 0.1% Tween for 2×10 min, and incubated with the corresponding secondary antibody for 30 min at room temperature. Chemiluminescence detection was performed with Amersham ECL (Cytiva, RPN2235; USA) and Bio-Rad Chemidoc XRS1 imaging system; Image Lab software v6.0.0 (Bio-Rad). Densitometry quantification analysis was done using Fiji software v1.53 [32]. The details of antibodies and their dilutions used for western blotting are described in ESM Table 5.
Flow cytometry
To quantify the percentage of definitive endoderm-positive cells at stage 1, flow cytometry for C-X-C motif chemokine receptor 4 was performed on one million dissociated cells using TrypLE (Thermo Fisher Scientific) for 3 min at 37°C. Cells were resuspended in cold 5% vol./vol. FBS-containing PBS and the conjugated antibodies were added and incubated for 30 min at room temperature. For intracellular antigen cytometry of stages 4, 5, 6 and 7, cells were dissociated with TrypLE for 8 min at 37°C and resuspended in cold 5% FBS-containing PBS. A total of one million cells were fixed and permeabilised using Cytofix/Cytoperm (554714, BD Biosciences, USA) as per manufacturer’s instructions. Primary, conjugated or IgG isotype (negative control) antibodies were incubated with the cells overnight at 4°C in Perm/Wash buffer (554714, BD Biosciences) containing 4% vol./vol. FBS and then secondary antibodies for 45 min at room temperature. The cells were then analysed using a FACSCalibur cytometer (BD Biosciences) with BD Cellquest Pro v4.0.2 (BD Biosciences) and FlowJo software v9 (BD Biosciences). The details of antibodies and their dilutions for flow cytometry are given in ESM Table 5.
Immunocytochemistry and immunohistochemistry
For whole mount or adherent culture staining, cells were fixed with 4% wt/vol. paraformaldehyde (PFA) for 15 min at room temperature, permeabilised with 0.5% vol./vol. Triton X-100 in PBS for 15 min at room temperature, then blocked with Ultra V block (Thermo Fisher Scientific) for 10 min followed by incubation with primary antibodies overnight at 4°C and secondary antibodies for 1 h at room temperature diluted in 0.1% vol./vol. Tween in PBS. For paraffin embedding, aggregates were fixed with 4% PFA at 4°C for 24 h then embedded in 2% low-melting agarose (Fisher Bioreagents; USA) PBS and transferred to paraffin blocks. Implanted grafts were retrieved after 3 months, dissected and fixed with 4% PFA at room temperature for 48 h before being paraffin embedded and cut into 5 µm sections using a Leica microtome. For immunohistochemistry, slides were deparaffinised and antigen-retrieved by boiling slides in 0.1 mol/l citrate buffer (pH 6) using a Decloaking chamber (Biocare Medical, USA) at 95°C for 12 min. For the TUNEL assay, an In Situ Cell Death Detection Kit, Fluorescein (no. 11684795910, Roche, Basel, Switzerland) was used according to the manufacturer’s instruction. EVOS FL Digital Inverted Fluorescence Microscope (Invitrogen, USA) or a Zeiss Axio Observer Z1 with Apotome (ZEN-2 software, Germany) were used for image acquisition. All samples were randomised, blinded, equally treated and acquired with the same microscope parameters. Image quantification was performed using CellProfiler software v4.2.1 [33], and Fiji software v1.53 [32]. The details of antibodies and their dilutions used in the study for immunofluorescence are described in ESM Table 5.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using a NucleoSpin Plus RNA kit (Macherey-Nagel, Germany). A total of 1.5 µg RNA was reversely transcribed using 0.5 μl Moloney murine leukaemia virus reverse transcriptase (Promega, M1701; USA), 4 µl RT buffer 5× (Promega), 2.5 μl dNTPs 2.5 mmol/l, 1 μl Oligo-dT 500 μg/ml (Promega), 0.2 μl Random hexamers 500 μg/ml (Promega) and 0.5 μl Riboblock RNase inhibitor 40 u/μl (Fermentas, USA) for 90 min at 37°C. The generated cDNA was amplified using 5× HOT FIREPol EvaGreen qPCR Mix Plus no ROX (Solisbiodyne, Estonia) in a 20 µl reaction. The reactions were pipetted using QIAgility (Qiagen, Germany) robot into a 100-well disc run in Rotor-Gene Q. Relative quantification of gene expression was analysed using the \({2}^{{-\Delta \Delta {\text{C}}}_{{\text{t}}}}\) method, with cyclophilin G (PPIG) as a reference gene. Reverse transcription without template was used as a negative control and an exogenous positive control was used as a calibrator. The RT-qPCR primers sequences are described in ESM Table 6.
Nonsense-mediated mRNA decay (NMD) cycloheximide inhibition assay and next-generation sequencing
At stage 3 of the differentiation protocol, heterozygous corrected HEL118.3 #RFX6+/− cells were treated with different concentrations (25, 50 and 100 µg/ml) of cycloheximide (Sigma-Aldrich, 01810; USA) for durations of 3 and 5 h. Total RNA was collected and reversely transcribed as described above but without using random hexamers. Primers with Illumina sequences attached were designed and used to amplify 120 bp in the variant region of the RFX6 cDNA, and 10 µl (5 ng/µl) were sequenced using next-generation sequencing (NGS) HiSeq PE100. The percentages of variant and corrected filtered reads from each of the alleles were calculated and plotted for the non-treated control and the cycloheximide treated samples. Primer sequences used for NGS cDNA amplification are described in ESM Table 1.
Ultra-deep bulk RNA-seq analysis
We performed ultra-deep RNA-seq analysis for H1 RFX6+/+ and RFX6−/− at stage 3 and stage 5, and for RFX6+/+ and RFX6+/− at stage 7 (SC-islets). Following mRNA library preparation using NEBNext Ultra II Directional RNA kit (New England Biolabs, Ipswich, USA), sequencing was performed using NovaSeq SP 2×100 bp v1.5 chemistry (Illumina, San Diego, USA). Preparation of RNA library and transcriptome sequencing was performed by the Sequencing laboratory of Institute for Molecular Medicine Finland FIMM Technology Centre, University of Helsinki. The raw data were filtered with cutadapt [34] to remove adapter sequences (with a minimum overlap of 5 bp), ambiguous (N) and low-quality bases (Phred score <25). We also excluded read pairs that were too short (<25 bp) after trimming. The filtered read pairs were mapped to the human reference genome (GRCh38) with STAR aligner [35]. Gene expression was counted from read pairs mapping to exons using featureCounts in Rsubreads [36], using GENCODE (GRCh38.p13) genome annotations [37]. Duplicate, chimeric and multimapping reads were excluded, as well as reads with low mapping scores (MAPQ <10). We obtained on average 31 million uniquely mapped read pairs per sample (between 27 million and 38 million).
We removed genes with very low or no expression, such as non-polyadenylated genes, from the analysis (<50 reads across all samples). The read count data were analysed with DESeq2 [38], comparing the expression differences between knockout and control samples in the different developmental stages. For stages 3 and 5 we compared the homozygous RFX6−/− variant with the control RFX6+/+ samples. Since RFX6−/− cells failed to differentiate into endocrine lineage, we compared the heterozygous RFX6+/− (clone 3G) to the control RFX6+/+ samples at stage 7 week 2. Each genotype per stage had four biological replicates. Genes were considered differentially expressed if the false discovery rate (FDR)-adjusted p values were <0.01. Principal component analysis (PCA) was calculated with prcomp (https://www.r-project.org/) using normalised log1p-transformed read counts. The differentially expressed genes (FDR<0.01) were analysed for enrichment separately for the up- and downregulated genes using clusterProfiler [39] against Reactome [40], KEGG [41], Disease ontology [42] and Gene ontology [43] databases. For heatmaps we used log1p-transformed normalised read counts, centred around gene mean using ComplexHeatmap [44]. For the Venn diagram we used ggvenn (https://github.com/yanlinlin82/ggvenn). The rest of the RNA-seq figures were made with ggplot2 [45]. Further details of the tools and software used for RNA-seq analysis are given in ESM Methods. The results from the gene expression analysis together with the raw sequences were deposited to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), with accession no. GSE234289.
Static and dynamic GSIS
For static GSIS assay, 50 aggregates were randomly picked and preincubated in 2.8 mmol/l glucose Krebs buffer in a 12-well plate placed on a rotating platform at 95 rev/min for 90 min at 37°C. Aggregates were then washed with Krebs buffer and sequentially incubated in Krebs buffer containing 2.8 mmol/l glucose, 16.8 mmol/l glucose and 2.8 mmol/l glucose plus 30 mmol/l KCl, on the rotating platform for periods of 30 min each. Samples of 200 µl were collected from each treatment and stored at −80°C for insulin ELISA measurements. Dynamic tests of insulin secretion were carried out using a perifusion apparatus (Suprafusion SF-06; Brandel, USA) with a flow rate of 0.25 ml/min and sampling every 4 min. Samples from each fraction collected were analysed using insulin ELISA (Mercodia, Uppsala, Sweden). Following static and dynamic tests of insulin secretion, the SC-islets were collected and the total insulin and DNA contents were analysed. Insulin content results were normalised to the DNA content of the beta cell fraction, calculated from flow cytometry insulin-positive cell percentage.
Electrophysiology
SC-islets were dispersed into single cells in cell dissociation buffer (Thermo Fisher Scientific) supplemented with trypsin (0.005% wt/vol.; Life Technologies, USA), washed and plated in serum-containing medium on 22 mm polylysine-coated coverslips, allowed to settle overnight, and then transduced with adenovirus coding for enhanced GFP under control of the RIP2 promoter to identify beta cells. Patch-clamp recordings were performed using an EPC-9 patch amplifier with PatchMaster v.2×90 software (HEKA Elektronik, Germany). Electrodes (resistance 2–4 MΩ) were pulled from borosilicate glass capillaries, coated with Sylgard and fire-polished. Cells were superfused with an extracellular solution containing (in mmol/l) 138 NaCl, 5.6 KCl, 1.2 MgCl2, 2.6 CaCl2, 10 glucose and 5 HEPES, pH 7.4 adjusted with NaOH at a rate of 0.4 ml/min at 32°C. Voltage-dependent currents and exocytosis were measured in whole-cell voltage-clamp mode with an intracellular solution containing (in mmol/l) 125 Cs-glutamate, 10 CsCl, 10 NaCl, 1 MgCl2, 0.05 EGTA, 3 Mg-ATP, 0.1 cAMP and 5 HEPES, pH 7.2 adjusted using CsOH. For current-voltage (IV) relationships, the membrane was depolarised from −70 mV to +80 mV (in steps of 10 mV lasting 50 ms each). Currents were compensated for capacitive transients and linear leak by subtracting the sum of the current responses to four depolarisations at 1/4 amplitude (P/4). Na+ and Ca2+ current components were separated by quantifying the initial peak current (0–5 ms; Na+) and average sustained current (5–45 ms; Ca2+). Exocytosis was quantified using the lockin module of Patchmaster (30 mV peak-to-peak; 1 kHz); with a train of 14×200 ms depolarisations to 0 mV at 1.4 Hz.
Exocytosis imaging
To visualise granule exocytosis, cells treated as described for electrophysiology were additionally infected with adNPY-tdOrange2 (a well-established marker for secretory granules) and imaged after 30–36 h using a custom-built lens-type total internal reflection fluorescence (TIRF) microscope based on an AxioObserver Z1 with a ×100/1.45 objective (Zeiss). Excitation was from two diode-pumped solid-state (DPSS) lasers at 491 and 561 nm (Cobolt, Sweden) passed through a cleanup filter (catalogue no. zet405/488/561/×640x; Chroma) and controlled with an acousto-optical tunable filter (AA-Opto). Excitation and emission light were separated using a beamsplitter (catalogue no. ZT405/488/561/640rpc; Chroma). The emission light was separated chromatically onto separate areas of an EMCCD camera (Roper QuantEM 512SC) using an image splitter (Optical Insights) with a cutoff at 565 nm (catalogue no. 565dcxr; Chroma) and emission filters (catalogue nos. ET525/50 m and 600/50 m, Chroma). Scaling was 160 nm per pixel. Cells were imaged in a standard solution containing 138 mmol/l NaCl, 5.6 mmol/l KCl, 1.2 mmol/l MgCl2, 2.6 mmol/l CaCl2, 10 mmol/l glucose, 5 mmol/l HEPES (pH 7.4 with NaOH), 200 μmol/l diazoxide (to block spontaneous depolarisation; Sigma-Aldrich), and 10 nmol/l exendin-4 (a glucagon-like peptide-1 receptor agonist; Anaspec). Exocytosis was evoked by rapidly depolarising cells with elevated K+ (75 mmol/l KCl equimolarly replacing NaCl in the standard solution, by computer-controlled local pressure ejection).
Cytoplasmic Ca2+ imaging
SC-islets were loaded with the fluorescent indicator Fura-2 LR (ion Biosciences) by incubation for 1 h with 1 μmol/l of its acetoxymethyl ester at 37°C in experimental buffer containing in (mmol/l) 138 NaCl, 4.8 KCl, 1.2 MgCl2, 2.56 CaCl2, 3 glucose and 25 HEPES (pH set to 7.40 with NaOH), and 0.5 mg/ml BSA. After rinsing in indicator-free buffer, the islets were attached to poly-l-lysine-coated coverslips in a 50 μl chamber on the stage of an Eclipse TE2000U microscope (Nikon) and superfused with buffer at a rate of 160 μl/min. The chamber holder and ×40, 1.3-NA objective were maintained at 37°C by custom-built thermostats. An LED light engine (LedHUB, Omicron Laserage Laserprodukte) equipped with 340 and 385 nm diodes and 340/26 nm (centre wavelength/half-bandwidth) and 386/23 nm interference filters (Semrock, IDEX Health & Science, LLC) provided excitation light that was led to the microscope via a liquid light guide. Emission was measured at 510/40 nm using a 400 nm dichroic beamsplitter and an Evolve 512 EMCCD camera (Photometrics). Image pairs at 340/386 nm were acquired every 2 s with the MetaFluor v.7.7 software (Molecular Devices). Cytoplasmic Ca2+ concentration ([Ca2+]i) was calculated from the background-corrected Fura-2 LR 340/380 nm fluorescence excitation ratio from manually defined cell-sized regions of interest. Large numbers of cells (>2000) were analysed at each condition, and differences between experimental groups are therefore unlikely to reflect different proportions of beta and non-beta cells. The data are presented as heatmaps from individual islets and cells and as scatter plots of the absolute [Ca2+]i values under different conditions. Calibration of Fura-2LR signal was performed to linearise the ratio signal using solutions without (0 Ca2+, 2 mmol/l EGTA) and with saturating Ca2+ (10 mmol/l) and 100 µmol/l Fura-2LR salt. The fluorescence ratio was converted to [Ca2+]i as described by Grynkiewicz et al [46].
In vivo animal implantation studies
Animal care and experiments were approved by the National Animal Experiment Board in Finland (ESAVI/14852/2018). NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NOD-SCID-Gamma [NSG], 005557, Jackson Laboratory; USA, https://www.jax.org/strain/005557) mice were obtained from SCANBUR and housed at Biomedicum Helsinki animal facility, on a 12 h light–dark cycle and fed standard chow ad libitum (2016 Teklad global 16% protein rodent diet, ENVIGO, USA). The temperature was kept at 23°C with 24% relative humidity. Implantations were performed on randomised 4- to 8-month-old male mice. Briefly, stage 6 RFX6−/−, and stage 7 RFX6+/+ and RFX6+/− SC-islets equivalent to 1.5 million cells (500 clusters) were loaded in PE-50 tubing and implanted under the kidney capsule. Mouse serum samples were collected monthly from the saphenous vein and stored at −80°C for human C-peptide analysis. Blood glucose levels were measured using ContourXT and ContourNext strips. Human-specific C-peptide was measured from plasma samples with the Ultrasensitive C-peptide ELISA kit (Mercodia, Uppsala, Sweden). To test the function of the SC-islet grafts, mice were subjected to an IPGTT. Before the test, mice were fasted 6 h, weighed and blood glucose was measured. Glucose (3 g/kg) was injected intraperitoneally, and blood samples (30 μl) were taken from the saphenous vein after 15, 30, 60 and 90 min to measure blood glucose and circulating human C-peptide levels by ELISA.
Quantification and statistical analysis
Data were collected from at least three independent differentiation experiments. Blinding was applied for immunohistochemical quantification. Morphological data represent population-wide observation from independent differentiation experiments. All representative images in the figures were reproducible in all the experiments performed and are followed by a quantification panel for all the experiments; the n value represents independent differentiation experiments, unless otherwise stated. Bar graphs are represented showing all the individual data points. Statistical methods used are described in each figure legend and individual method section. The results are presented as the mean ± SD unless otherwise stated. p values<0.05 were considered statistically significant.
Results
Impact of heterozygous and homozygous RFX6 frameshift variant p.His293LeufsTer7 on diabetes development
A two-nucleotide deletion of the RFX6 gene in exon 9 (c.878_879delAC) leads to a frame shift at codon 293 and an early stop codon at 298 (p.His293LeufsTer7), instead of the 928 amino acid wild-type (WT) protein [47]. While the WT protein contains a winged-helix RFX6 DNA binding domain and three dimerisation domains mediating both hetero- and homodimeric interactions [6], this frameshift variant is predicted to contain only the DNA binding domain and the first dimerisation domain (Fig. 1a). However, in silico protein localisation prediction tools identified the loss of a nuclear localisation sequence, resulting in a predicted cytoplasmic localisation, which is unlikely to interfere with the DNA binding of the WT protein (data not shown) [48,49,50].
Impact of heterozygous and homozygous RFX6 frameshift variant p.His293LeufsTer7 on diabetes development. (a) The human RFX6 gene and the encoded WT and predicted mutant proteins. The WT RFX6 is a 928-amino-acid protein containing a DNA binding domain (DBD) and three dimerisation domains (B, C and Dimerisation). The RFX6 frameshift variant p.His293LeufsTer7 contains the DBD, dimerisation domain B and a frameshift making an early stop codon at 298. The antibody used in this study binds to the amino acid sequence from 676–762 (shown in yellow). (b) Survival curve stratified by RFX6 genotype (WT, n=439,416 and Het; heterozygous carriers, n=1318) and adjusted HR for type 2 diabetes risk. The mean ± SD age at onset for Het and WT was 57.48±12.87 and 59.91±12.43 years, respectively (p=2.5×10−4). The total number of individuals in the FinnGen dataset was 440,734, which included 71,728 with type 2 diabetes. (c) Survival curve stratified by RFX6 genotype (WT, n=253,803 and Het; heterozygous carriers, n=814) and adjusted HR for gestational diabetes risk. The total number of individuals in the FinnGen dataset was 254,617, which included 16,802 with gestational diabetes. (d) Schematic showing the patient with Mitchell–Riley syndrome’s clinical manifestations (pancreatic hypoplasia with neonatal diabetes, gall bladder agenesis and intestinal stenosis with malabsorptive diarrhoea). (e) Schematic of generating RFX6 allelic series of the frameshift variant in H1 hESCs and patient-derived iPSCs, followed by directed differentiation into pancreatic endocrine cells. Panel (e) was created using Servier Medical Art. Survival plots (b, c) were generated using survminer and adjusted HRs were calculated using Cox proportional hazards model adjusting for age, sex and principal components PC1–PC10
We assessed the impact of this heterozygous frameshift variant on the lifetime risk of type 2 and gestational diabetes in the FinnGen dataset (data freeze R11), containing data from 440,734 individuals. The 1,318 heterozygous carriers of the variant showed 80% higher risk of type 2 diabetes (p=6.8×10−28), with, on average, a 2 year earlier age of onset (Fig. 1b), and 79% higher risk of gestational diabetes (p=2.3×10−8) (Fig. 1c). Interestingly, the carriers also demonstrated a reduced BMI (β=−0.12 SD, p=1.3×10−4 and n=321,672) compared with non-carriers.
We identified one male individual carrying this frameshift variant in homozygous form. This boy is currently alive at 10 years of age. He was diagnosed with Mitchell–Riley syndrome at birth, manifesting pancreatic hypoplasia, neonatal diabetes, gall bladder agenesis, intestinal stenosis and malabsorptive diarrhoea (Fig. 1d). The individual had low birth size, which recovered gradually after administering total parenteral nutrition (ESM Fig. 1a). At the age of 4 years, magnetic resonance cholangiopancreatography showed a hypoplastic pancreas of 4 ml (data not shown); the average volume is 20 ml for healthy individuals of the same age [51]. Blood glucose was normal at birth but reached hyperglycaemic levels at the age of 4 days. Insulin treatment was started at a daily dose of 0.1 U/kg, as C-peptide was barely detectable (ESM Fig. 1b). Plasma glucagon was undetectable at birth but, curiously, it normalised gradually over the course of 4 months (ESM Fig. 1c). In contrast, PP dropped below the detection limit (ESM Fig. 1d). Pancreatic exocrine function displayed severe impairment (ESM Fig. 1e). Considering the characteristic clinical features of this homozygous frameshift variant aligning with typical Mitchell–Riley syndrome, and the predicted loss of nuclear localisation of the protein, we have denoted this variant as a loss-of-function variant RFX6−/−.
To study the p.His293LeufsTer7 variant in more detail, we generated two isogenic allelic series stem cell models. In the first model, we introduced the variant in H1 hESCs. We validated two heterozygous clones (RFX6+/− 9C and RFX6+/− 3G) and one homozygous (RFX6−/−) clone (Fig. 1e and ESM Fig. 2). In the second model, we generated patient-derived iPSCs from the homozygous patient and then corrected the variant heterozygously and homozygously, creating the iPSC lines #RFX6−/−, #RFX6+/− and #RFX6+/+ (Fig. 1e and ESM Fig. 3).
RFX6 controls the transcriptional network of pancreatic development
We differentiated the generated hESCs towards functional SC-islets using our optimised seven-stage protocol [28, 29] (Fig. 2a). Since RFX6 expression started to increase at the posterior foregut stage (S3) (Fig. 2b), the frameshift variant did not affect the earlier stage of definitive endoderm induction (data not shown). However, levels of RFX6 gene expression in both heterozygous RFX6+/− and homozygous RFX6−/− cells were reduced (Fig. 2b). The loss of RFX6 in RFX6−/− cells was confirmed by immunoblotting at S3 and, interestingly, both heterozygous clones showed significant reduction in RFX6 protein levels at this stage, indicating haploinsufficiency (Fig. 2c,d). The lack of RFX6 in RFX6−/− cells was associated with a significant reduction in expression of PDX1, a key transcription factor in pancreatic development (Fig. 2e). Subsequently, the percentage of PDX1+ NKX6.1+ and the emergence of cells positive for chromogranin A (CHGA) were significantly reduced (markers of pancreatic endocrine lineages), while both heterozygous clones were similar to RFX6+/+ (Fig. 2f and ESM Fig. 4).
RFX6 controls the transcriptional network of pancreatic development. (a) Schematic of H1 SC-islet differentiation protocol. Stages 1–4 in monolayer, stage 5 in microwells and stages 6–7 in suspension culture, followed by implantation under kidney capsule of NSG mice. (b) Relative gene expression levels of RFX6 at stage 0 (S0), S3, S5 and S6 (n=3–5). (c) Protein immunoblots of RFX6 and β-actin for the isogenic H1 clones (9C and 3G) at S3. (d) Percentage of RFX6 protein band densitometry normalised to β-actin bands, quantified from (c) (n=3 or 4). (e) Relative gene expression levels of PDX1 at S0, S3, S5 and S6 (n=3–5). (f) Percentage of PDX1+ and NKX6.1+ cells at S5 measured by flow cytometry (n=6–10). (g) Heatmap showing the relative differences in gene expression of RFX6+/+ and RFX6−/− at S3 and S5 (n=4). Using log1p transformed normalised read count, with sample specific expression subtracted from gene averages. Each gene is differentially expressed in one or both of the stages, FDR<0.01. (h) Enrichment analysis of the differentially expressed genes between RFX6+/+ and RFX6−/− at S3 and S5. The upregulated and downregulated genes (FDR<0.01) were analysed for enrichment against the Reactome database separately for S3 and S5 comparisons. Statistical significance was measured using two-way ANOVA with Tukey’s test for multiple comparisons correction in (b, e), and one-way ANOVA with Tukey’s test for multiple comparisons correction in (d, f). Data are presented as means ± SD; ***p<0.001
Since the quantity and quality of pancreatic progenitors and endocrine precursors were impaired in RFX6−/−, but not in RFX6+/− cells, we ran bulk RNA-seq analysis to identify differentially expressed genes between RFX6−/− and RFX6+/+ at stages 3 and 5 (S3 and S5; ESM Fig. 5). In RFX6−/− cells, the transcription factor regulatory network orchestrating the development of the pancreatic lineage was downregulated at S3 including RFX6, PDX1, NKX6.1 (also known as NKX6-1), FEV, PAX4, NEUROG3 and NEUROD1 (Fig. 2g). While most of these genes remained downregulated at S5 in addition to ISL1 and PAX6, expression levels of NEUROG3 and NKX2.2 recovered, with a significant increase in PAX4 compared with RFX6+/+. Interestingly, expression levels of all pancreatic hormones were downregulated at S5, except for PPY, which was significantly upregulated (Fig. 2g and ESM Fig. 6a). In agreement with these findings, Reactome enrichment analysis showed downregulation of pathways involved in beta cell development and gene expression in RFX6−/− cells. Additionally, voltage-gated potassium channels were reduced at S3, and the regulation of insulin secretion, and glucose-dependent insulinotropic polypeptide (GIP) synthesis and secretion were reduced at S5 (Fig. 2h).
We further confirmed these findings in our isogenic iPSC model using a modified version of a simplified monolayer differentiation protocol [31]. As observed with hESCs, the emergence of cells positive for pancreatic and duodenal homeobox 1 (PDX1) in #RFX6−/− was not compromised, while RFX6 and PDX1 gene expression and the generation of NK6 homeobox 1 (NKX6.1)-positive pancreatic progenitors were significantly reduced (ESM Fig. 7 and ESM Fig. 8a, b). We then investigated if the cells carrying the frameshift variant exhibit reduced RFX6 expression due to NMD. We treated #RFX6+/− cells with the NMD inhibitor cycloheximide at S3 and quantified the reads of variant and corrected transcripts. In the non-treated samples, the percentage of frameshift variant cDNA was only 15%, while treatment with cycloheximide gradually normalised the expression to ~48% in a dose-dependent manner, confirming that the frameshift variant mRNA was subjected to NMD (ESM Fig. 8c). At the endocrine stage (S6), #RFX6−/− cells lacked the expression of the five pancreatic hormones, insulin, glucagon, somatostatin, PP and ghrelin, while this phenotype was rescued in both the heterozygous and homozygous corrected cells (ESM Fig. 8d, e).
Persistent expression of SRY-box transcription factor 9 and neurogenin 3 in homozygous RFX6 −/− cells
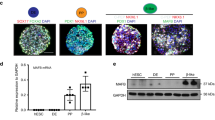
We further studied the endocrine precursors in the 3D model of hESCs and found a peculiar dysregulation of SRY-box transcription factor 9 (SOX9) and neurogenin 3 (NEUROG3). Normally, when SOX9 is highly expressed in the pancreatic epithelium it induces the proendocrine transcription factor NEUROG3, which upon reduction of notch signalling promotes the endocrine lineage differentiation program [52]. Indeed, in our S4 pancreatic progenitors, SOX9 was highly expressed across all lines (Fig. 3b and ESM Fig. 4a). Upregulation of NEUROG3 expression at S5 was associated with downregulation of SOX9 expression (Fig. 3a–c). A further decrease in SOX9 expression was noticed in the immature S6 SC-islets in RFX6+/+ and RFX6+/− but not in RFX6−/− cells (Fig. 3b). Interestingly, NEUROG3 expression increased at S5 in RFX6−/− cells, similar to the observation in bulk RNA-seq, but the difference did not reach statistical significance. However, it was significantly higher than in the other genotypes at S6 (Fig. 3c). The pattern of SOX9 and NEUROG3 expression was similar between the genotypes in S5 endocrine precursors, as shown by immunohistochemistry (Fig. 3a). However, the pattern was completely altered at S6; whereas SOX9 was detected mainly in the cytoplasm of RFX6+/+ and RFX6+/− cells, nuclear SOX9 expression persisted in RFX6−/− cells (Fig. 3d). The numbers of nuclear SOX9+ cells and NEUROG3+ cells were significantly increased in RFX6−/− cells (Fig. 3e,f). Since NEUROG3 expression induces the endocrine lineage specification, we investigated the expression of the endocrine marker CHGA. While ~80% of the cells were CHGA+ in RFX6+/+ and RFX6+/− cells, only ~43% were CHGA+ in RFX6−/− cells, with less than 1% being insulin-positive (Fig. 3g,h and ESM Fig. 9a). Most CHGA+ cells in RFX6−/− cells displayed positivity for solute carrier family 18 member A1 (SLC18A1), a marker of enterochromaffin cells (a subpopulation commonly observed in various differentiation protocols, including ours [28, 53, 54]), although they were disproportionately increased in RFX6−/− (ESM Fig. 9b). A significant increase in apoptosis was detected in RFX6−/− cells by TUNEL assay at the end of S6 (Fig. 3i,j), and the cells did not survive for more than 2 days at the final maturation stage 7 (S7). This suggests that the absence of RFX6, and consequently the failure to specify pancreatic endocrine precursors, results in progressive cell death.
Persistent expression of SOX9 and NEUROG3 in homozygous RFX6−/− cells and increased apoptosis. (a) Immunohistochemistry showing SOX9+ and NEUROG3+ cells at stage 5 (S5). Scale bar, 50 µm. (b, c) Relative gene expression levels of SOX9 (b) and NEUROG3 (c), comparing all the cell lines at S4, S5 and S6 (n=4–9). (d) Immunohistochemistry for SOX9+ and NEUROG3+ cells at S6. Scale bar, 50 µm. (e, f) Percentages of SOX9+ (e) and NEUROG3+ cells (f) at S6 quantified from (d) (n=4–5). (g, h) Percentage of CHGA+ (g) and insulin (INS)+ cells (h) at S6 quantified by flow cytometry (n=5–7). (i) Immunohistochemistry showing TUNEL+ and CHGA+ cells for the RFX6+/+ and RFX6−/− clones at S6. Scale bar, 50 µm. (j) Percentage of TUNEL+ cells at S6 quantified from (i) (n=3–4). Statistical significance was measured using two-way ANOVA with Tukey’s test for multiple comparisons correction in (b, c), one-way ANOVA with Tukey’s test for multiple comparisons correction in (e–h) and two-tailed unpaired t test in (j). Data are presented as means ± SD; *p<0.05, **p<0.01, ***p<0.001
RFX6 haploinsufficiency impairs insulin secretion of beta cells
Quantifying all endocrine populations (positive for insulin, glucagon, somatostatin, PP, ghrelin and SLC18A1), we verified an extremely limited presence of pancreatic hormone cells in RFX6−/− cells, with no significant difference between RFX6+/+ and RFX6+/− cells. Although there was a slight increase in enterochromaffin cells (SLC18A1+) in RFX6+/−, the difference did not reach statistical significance (ESM Fig. 10).
Since RFX6−/− cells failed to survive at S7, we continued to study the function of only the WT and heterozygous cells. The proportions of insulin- and glucagon-positive cells (markers of beta cells and alpha cells, respectively) and total insulin content were similar between the two genotypes (Fig. 4a–d and ESM Fig. 11a). We confirmed the persistent RFX6 haploinsufficiency at this stage using protein immunoblotting, showing 54.2% reduction in protein expression (Fig. 4e,f). GSIS in a static assay, normalised to DNA content, showed decreased insulin secretion in RFX6+/− compared with WT cells in both basal and high glucose conditions (62% and 54% reduction, respectively), while KCl-stimulated insulin secretion showed no difference (Fig. 4g). However, glucose stimulatory index was similar between RFX6+/+ and RFX6+/− cells (ESM Fig. 11b). Correspondingly, dynamic perifusion assays confirmed the reduced insulin secretion in both glucose conditions (Fig. 4h). They also revealed a blunted first-phase response, as well as lower insulin secretion when stimulated with the glucagon-like peptide 1 (GLP-1) analogue exendin-4. Insulin secretion shut down effectively when the glucose concentration was decreased back to 2.8 mmol/l in both genotypes, followed by a normal maximum secretion capacity with KCl depolarisation (Fig. 4h). Total AUC for the entire perifusion assay was significantly reduced in RFX6+/− cells, including the individual phases of low glucose, high glucose, and high glucose with exendin-4 (Fig. 4i–n). These findings highlight that lower RFX6 expression in the heterozygous cells reduces the insulin secretion capacity in basal and stimulatory glucose conditions without impacting beta cell number or insulin content.
RFX6 haploinsufficiency impairs insulin secretion of beta cells. (a) Immunohistochemistry showing insulin-positive (INS+) and glucagon-positive (GCG+) cells for RFX6+/+ and RFX6+/− SC-islets at stage 7 week 2 (S7w2), and for RFX6−/− at stage 7 day 2 (S7d2). Scale bar, 50 µm. (b, c) Percentage of INS+ (b) and GCG+ cells (c) for RFX6+/+ and RFX6+/− SC-islets at S7w2 measured by flow cytometry (n=7–9). (d) Insulin content of RFX6+/+ and RFX6+/− SC-islets at stage 7 week 3 (S7w3) normalised to the DNA content of beta cells (n=7 or 8). (e) Protein immunoblots of RFX6 and β-actin for RFX6+/+ and RFX6+/− SC-islets at S7w2. (f) Percentage of RFX6 protein bands densitometry normalised to β-actin bands, quantified from (e) (n=3 or 4). (g) Static insulin secretion at S7w3 at low 2.8 mmol/l glucose (G2.8), followed by high 16.8 mmol/l glucose (G16.8) and then depolarisation with G2.8+30 mmol/l KCl, normalised to the DNA content of the SC-islets (n=7–9). (h) Dynamic insulin secretion responses to perifusion (4 min intervals) with G2.8, G16.8, G16.8+12 nmol/l exendin-4 (Ex-4) and G2.8+30 mmol/l KCl, normalised to the DNA content of the SC-islets (n=7 or 8). (i) Total AUC, quantified from (h) (n=7 or 8). (j–n) AUC for individual phases, quantified from (h) (n=7 or 8). Statistical significance was measured using unpaired t test in (b, c, i–n), using one-way ANOVA with Tukey’s test for multiple comparisons correction in (d, f) and using two-tailed unpaired multiple t tests in (g, h). Data are presented as means ± SD; *p<0.05, **p<0.01, ***p<0.001
Reduced [Ca2+]i in basal and depolarised conditions in RFX6 +/− SC-islets
To mechanistically understand the reduced glucose sensitivity of RFX6+/− SC-islets, we explored the stimulus–secretion coupling machinery of SC-islet beta cells by analysing [Ca2+]i, ion channel conductance and exocytosis. The [Ca2+]i was recorded from individual cells in SC-islets loaded with Fura-2LR. In line with reduced basal insulin secretion, [Ca2+]i at 3 mmol/l glucose was significantly lower in RFX6+/− than in RFX6+/+ SC-islets (Fig. 5a,b). Most of the SC-islet cells responded to 16.7 mmol/l glucose with [Ca2+]i elevation, indicating beta cell identity. The glucose-induced [Ca2+]i increase was moderately reduced in RFX6+/− SC-islets but the difference did not reach statistical significance. Blockade of KATP channels with 1 mmol/l tolbutamide increased [Ca2+]i to an extent comparable with glucose stimulation. Depolarisation with 30 mmol/l K+ resulted in a more pronounced [Ca2+]i increase, which was significantly reduced in RFX6+/− compared with RFX6+/+ SC-islets (Fig. 5a,b). Patch-clamp recordings showed no difference in amplitude or voltage-dependence of total and nifedipine-sensitive Ca2+ currents or of Na+ currents (ESM Fig. 11c, d). Exocytosis was measured in single cells as changes in membrane capacitance. The pre-stimulatory capacitance was similar in control and RFX6+/− cells, indicating no difference in cell size (ESM Fig. 11e). A train of depolarisations (14×200 ms) resulted in slightly larger capacitance increase in RFX6+/− cells (ESM Fig. 11f, g) but the difference was not statistically significant. Exocytosis was also measured by TIRF microscopy in cells expressing the granule marker NPY-tdmOrange2. Control and RFX6+/− cells showed similar granule density at the plasma membrane and comparable degree of granule fusion events following depolarisation with high K+ combined with diazoxide (ESM Fig. 11h, i). Together, these experiments indicate that RFX6+/− SC-islets have no general impairment of exocytosis capacity and that the reduced GSIS is most likely explained by other factors, such as altered generation of metabolic coupling factors that amplify Ca2+-triggered exocytosis.
To further investigate the findings of lower insulin secretion and reduced [Ca2+]i, we performed transcriptome analysis at S7 using bulk RNA-seq (ESM Fig. 5). The gene expression levels of RFX6 were downregulated in the heterozygous cells, in line with the observed lower protein expression and functional haploinsufficiency. Concomitantly, beta cell maturation markers, such as G6PC2 [28], UCN3 [55], ADCY7 [56] and PRKCA [57,58,59], together with voltage-gated calcium and potassium channels, were reduced compared with expression in RFX6+/+ cells (Fig. 5c and ESM Fig. 12a). In contrast, pancreatic progenitor genes such as, ONECUT3 [60], NKX6-3 [61, 62] and HNF1B [63] were upregulated (Fig. 5d and ESM Fig. 12b), indicating an immature phenotype of the heterozygous SC-islets. Notably, the serotonin biosynthesis gene TPH1 was significantly increased, aligning with the observed, albeit non-significant, rise in enterochromaffin cells (ESM Fig. 10e), and consistent with findings in Rfx6-deficient mice [64]. Moreover, upregulation of the disallowed genes IGF2 [65], CACNB3 [66, 67] and KCNQ1 [68, 69] was detected (Fig. 5d and ESM Fig. 12b). However, the gene expression of pancreatic hormones was unaltered (Fig. 5e and ESM Fig. 12c). These findings illustrate the immature phenotype of RFX6+/− SC-islets, with reduced [Ca2+]i and insulin secretion.
Reduced [Ca2+]i in basal and depolarised conditions in RFX6+/− SC-islets. (a) Heatmaps of [Ca2+]i recorded with Fura-2 LR in cells within SC-islets under basal conditions of 3 mmol/l glucose (G3) and after stimulation with 16.7 mmol/l glucose (G16.7), G3+1 mmol/l tolbutamide (tolb) and G3+30 mmol/l K+ (K+30). Each heatmap shows the responses of all cells in one islet. (b) Quantification of [Ca2+]i responses from experiments as in (a). Time-averaged [Ca2+]i for all individual cells (dots) and average values for all cells in each islet (circles). The bars represent means ± SEM for the averaged islet responses. n=no. of islets (total cell number in parenthesis): G3 WT, n=65 (6933); G3 RFX6+/−, n=75 (8502); G16.7 WT, n=29 (3203); G16.7 RFX6+/−, n=42 (4853); tolb WT, n=23 (2720); tolb RFX6+/−, n=39 (4692); K+30 WT n=32 (3639); and K+30 RFX6+/−, n=37 (4320). Statistical analyses with Student’s unpaired two-tailed t test, ***p<0.001. (c) Heatmap of downregulated genes in RFX6+/− SC-islets at stage 7 week 2 (S7w2). Statistically significant genes (ADCY7, G6PC2, UCN3, KCTD12, KCNIP3 and CACNA1B), FDR<0.01 (n=4). (d) Heatmap of upregulated genes in RFX6+/− SC-islets at S7w2. All genes except IGFBP3 are statistically significant, FDR<0.01 (n=4). (e) Heatmap of hormone gene expression in RFX6+/+ and RFX6+/− SC-islets at S7w2. None of the genes show statistically significant differences (n=4). Heatmaps (c–e) show log1p transformed normalised read count, with sample-specific expression subtracted from gene averages
Heterozygous RFX6 frameshift variant beta cells present lower insulin secretion in vivo
To investigate whether the reduced insulin response persisted after further maturation of the SC-islets in vivo, we implanted 500 S7 RFX6+/+ or RFX6+/− SC-islets under the kidney capsule of immunocompromised non-diabetic NSG mice. Additionally, we implanted S6 RFX6−/− SC-islets to evaluate the fate of the surviving cells in vivo, since they failed to survive in vitro. Blood glucose and human C-peptide levels were measured monthly for 3 months, followed by an IPGTT (Fig. 6a). The normal range of blood glucose levels in random-fed NSG mice is between 6 and 8.5 mmol/l; however, starting from the second month post implantation, the blood glucose levels of the mice implanted with RFX6+/+ SC-islets dropped to the human level of ~4.6 mmol/l (Fig. 6b). In contrast, mice implanted with RFX6+/− SC-islets required an additional month to display human blood glucose levels (Fig. 6b). RFX6−/− cells failed to reduce the mouse blood glucose levels. While human C-peptide levels were undetectable in mice implanted with RFX6−/− SC-islets, they increased steadily in the mice implanted with RFX6+/+ and RFX6+/− SC-islets. However, throughout the 3 months, the C-peptide levels in mice implanted with RFX6+/− SC-islets were approximately half those in their RFX6+/+-implanted counterparts (Fig. 6c).
Heterozygous RFX6+/− beta cells present lower insulin secretion in vivo. (a) Schematic showing the data points and sampling post implantation of SC-islets under the kidney capsule of NSG mice. (b) Random blood glucose levels measured monthly post implantation (n=3–11). (c) Random human C-peptide levels measured monthly post implantation (n=3–8). (d) Immunohistochemistry showing insulin-positive (INS+) and glucagon-positive (GCG+) cells at month 3 post implantation. In RFX6−/− cells, yellow arrows point to INS+ and white arrows point to GCG+. Scale bar, 200 µm. (e) Immunohistochemistry showing CHGA+ and PDX1+ cells at month 3 post implantation. Scale bar, 200 µm. (f) Fasting blood glucose levels measured at month 3 post implantation (n=7 or 8). (g) Fasting human C-peptide levels measured at month 3 post implantation (n=3–8). (h) Blood glucose levels during an IPGTT of fasted mice at month 3 post implantation (n=7–9). (i) AUC quantified from (h) (n=7–9). (j) Human C-peptide during an IPGTT of fasted mice at month 3 post implantation (n=2–7). (k) AUC quantified from (j) (n=2–7). Statistical significance was measured using two-way ANOVA with Tukey’s test for multiple comparisons correction in (b, c), one-way ANOVA with Tukey’s test for multiple comparisons correction in (f–i) and two-tailed unpaired t test between RFX6+/+ and RFX6+/− in (j, k). Data are presented as means ± SD, except for (h, j) showing mean ± SEM; *p<0.05, **p<0.01, ***p<0.001
Immunohistochemical analysis after 3 months showed abundant insulin-positive and glucagon-positive cells in RFX6+/+ and RFX6+/− grafts, while these cell types were rarely found in RFX6−/− implants (Fig. 6d). Similarly, most of the implanted RFX6+/+ and RFX6+/− cells were CHGA-positive, with an abundance of PDX1-positive cells, representing insulin-positive cells (Fig. 6e). In contrast, most cells were negative for both markers in RFX6−/− implants and the morphology of cells was distinctly different. To evaluate the origin of these cells, we performed immunohistochemical analysis using a human-specific mitochondrial antibody (hMITO), together with SOX9 immunostaining, that persists in RFX6−/− in vitro. Indeed, insulin-positive cells from the RFX6+/+ implants were positive for hMITO, while the surrounding cells from the mouse tissue were negative (ESM Fig. 13a). The morphologically different cells in the RFX6−/− implants were positive for hMITO and nuclear SOX9, compared with scarce cytoplasmic SOX9-positive cells in RFX6+/+ implants. Both RFX6+/+ and RFX6−/− cells were negative for NEUROG3 expression at 3 months post implantation (ESM Fig. 13b). Since PP cells were found in Rfx6−/− mice, we stained for PP and did not detect any PP-positive cells in the RFX6−/− grafts (ESM Fig. 13c). This suggests that RFX6 is essential for the development of all pancreatic endocrine cells in humans, including PP cells.
At 3 months post implantation, RFX6+/+ and RFX6+/− implanted mice had a blood glucose level of ~3.9 mmol/l, similar to the fasting level in humans, while non-implanted mice presented the normal mouse blood glucose levels of ~7 mmol/l (Fig. 6f). However, human C-peptide levels in fasting mice implanted with RFX6+/− SC-islets were ~60% lower (Fig. 6g). In the IPGTT, both RFX6+/+ and RFX6+/− SC-islets were able to control the abrupt increase in blood glucose levels similarly, and more efficiently, compared with the non-implanted mice, which had higher blood glucose levels at each of the measured time points (Fig. 6h) and in the measured AUC (Fig. 6i). Although the secreted human C-peptide levels of both genotypes had a similar pattern and peaked at 30 min post injection, the levels were lower in the RFX6+/− implanted mice (Fig. 6j) and the total secretion AUC was significantly reduced (Fig. 6k). These findings indicate that RFX6 haploinsufficiency due to the lower gene dose in RFX6+/− SC-islets leads to persistent reduction in insulin secretion.
Discussion
A hierarchy of gene regulatory networks control the development and function of beta cells. Single pathogenic variants in more than 20 transcription factors of this network can lead to diabetes. Importantly, species differences in the developmental and functional processes of beta cells impede their precise study with mouse models. Here, we sought to overcome this limitation using genetically engineered human SC-islets, to investigate the developmental and functional defects of beta cells upon RFX6 perturbation.
Our findings demonstrate that RFX6 p.His293LeufsTer7 is a loss-of-function variant with a transcript subjected to nonsense-mediated decay. We confirmed that the lower RFX6 gene dose in heterozygous RFX6 SC-islets led to RFX6 haploinsufficiency, which did not compromise the differentiation capacity of the cells in terms of beta cell number or insulin content, but impaired beta cell function. There is no association between Rfx6 haploinsufficiency and diabetes in mice, indicating that this phenotype cannot be faithfully modelled in rodents [6]. Similar discrepancies between diabetes development in humans and mice have been reported with variants in HNF1A, HNF1B and PAX4. While pathogenic heterozygous variants in all of these genes cause MODY in humans, heterozygous null mice lack any diabetes phenotype [70,71,72]. However, our findings are in line with the beta cell-specific Rfx6 knockout in adult mice [12] and confirm the reported reduction of insulin secretion in islets from donors with type 2 diabetes as a result of reduced RFX6 levels [14]. Reduced RFX6 expression in RFX6+/− SC-islets led to decreased levels of the beta cell marker UCN3 and VDCC expression, lower [Ca2+]i and reduced insulin secretion in basal and high glucose levels. Moreover, RFX6+/− SC-islets showed upregulation of disallowed genes that are reported to negatively impact insulin secretion, such as IGF2 [65], CACNB3 [66, 67] and KCNQ1 [68, 69].
Insulin content of the heterozygous cells was not affected, suggesting that the difference in insulin secretion more likely reflects impaired stimulus–secretion coupling or granule trafficking. However, exocytosis measured either as changes in capacitance or by TIRF imaging of granules at the plasma membrane was not reduced in response to depolarising stimuli. This is in line with intact K+-induced insulin secretion in static and dynamic perifusion experiments. Despite slightly reduced gene expression levels of some voltage-gated Ca2+ channel components, there was no difference in Ca2+ currents, unlike the reduced Ca2+-channel activity that was reported in RFX6 knockdown in the EndoC-βH2 cell line [13]. The reduced basal [Ca2+]i and the lower [Ca2+]i after membrane depolarisation may instead reflect differences in Ca2+ transport or buffering. The lower basal [Ca2+]i is consistent with reduced insulin secretion at low glucose. Since the substimulatory [Ca2+]i influences granule priming [73], reduced resting [Ca2+]i may indirectly impair secretion also at elevated glucose. However, the intact K+-induced insulin secretion, and the reduced GSIS despite insignificantly altered [Ca2+]i, more likely point towards a role for RFX6 in controlling the metabolic amplifying pathway. The [Ca2+]i elevation triggered by K+ depolarisation was significantly reduced in RFX6+/− cells, yet Ca2+ influx near insulin granules might still reach a level sufficient to induce a maximum insulin secretion response, as evidenced in both static and dynamic insulin secretion assays. Based on these observations, we suggest insulin secretagogues and/or GLP-1 receptor agonists as treatment to enhance the secretory capacity of beta cells in heterozygous RFX6 variant carriers.
The heterozygous RFX6 SC-islets sustained lower insulin secretion in vivo, resulting in delayed lowering of blood glucose post implantation. All of our findings from the heterozygous cell model are consistent with the increased risk of variant carriers to develop diabetes, as shown for both gestational and type 2 diabetes, and previously for MODY with reduced penetrance [15]. Further exploration of the clinical phenotype in variant carriers is warranted. However, two small studies have indicated elevated fasting glucose [15] or 2 h glucose [47] during an OGTT in carriers compared with non-carriers. Additionally, lower fasting and stimulated GIP levels were observed [15]. Consistent with an impact on insulin secretion, carriers displayed a younger age at diagnosis of diabetes and a lower BMI.
The complete loss of RFX6 led to reduced expression of PDX1, NKX6.1, FEV, ISL1, NEUROD1 and PAX6 with gradual loss of pancreatic progenitor identity. These genes are in line with some of the reported targets of ChIP-seq analysis of whole pancreatic tissue from adult mice (e.g. Pdx1, Neurod1 and Nkx6.1) [74]. Although, the pancreatic markers NEUROG3, NKX2.2 and PAX4 were reduced at an earlier stage of the differentiation, they recovered at the endocrine precursor stage. This phenotype is similar to that reported in Rfx6 knockout mice [6, 12], with a difference of reduced NKX6.1 expression in our model. While gene expression levels of PPY were increased in RFX6−/− endocrine precursors, PP cells formed in extremely limited numbers in vitro and were not found after 3 months following implantation in vivo, recapitulating the loss of pancreatic polypeptide in the plasma of the Mitchell–Riley syndrome patient, in contrast to the increased numbers of PP cells in the Rfx6−/− mouse model [6]. We demonstrate persistent expression of NEUROG3 and SOX9 at the pancreatic endocrine stage in RFX6−/− cells, suggesting a negative regulatory role of RFX6 on SOX9 and NEUROG3 expression. Most of the emerging endocrine cells exhibited enterochromaffin cell identity with a pancreatic endocrine specification barricade, followed by increased cell death upon further differentiation in vitro. Reduced generation of the pancreatic progenitor pool and increased apoptosis phenotype could explain the pancreatic hypoplasia seen in homozygous individuals. Implantation of the homozygous cells confirmed persistent SOX9 expression in the majority of the grafted cells, while CHGA-expressing cells were extremely rare. This suggests that RFX6 functions to limit SOX9 expression to allow full endocrine differentiation. Human C-peptide was not detected in mice implanted with RFX6−/− cells. The presence of small numbers of insulin-positive and glucagon-positive cells may explain the very low C-peptide levels observed in the patient but not the normal glucagon levels. These normal glucagon levels might be explained by potential extrapancreatic secretion of the hormone [75].
In summary, we highlight the critical role of RFX6 in augmenting and maintaining the pancreatic progenitor pool, with an endocrine roadblock upon its loss, persistent NEUROG3 and SOX9 expression and increased cell death. We demonstrate that RFX6 haploinsufficiency does not affect beta cell number or insulin content but does impair function, predisposing carriers to diabetes. Our allelic series isogenic SC-islet models represent a powerful tool to elucidate specific aetiologies of diabetes in humans, enabling the sensitive detection of aberrations in both beta cell development and function.
Abbreviations
- [Ca2+]i :
-
Cytoplasmic Ca2+ concentration
- CHGA:
-
Chromogranin A
- FDR:
-
False discovery rate
- GIP:
-
Glucose-dependent insulinotropic polypeptide
- GLP-1:
-
Glucagon-like peptide 1
- GSIS:
-
Glucose-stimulated insulin secretion
- hESC:
-
Human embryonic stem cell
- hMITO:
-
Human-specific mitochondrial antibody
- iPSC:
-
Induced pluripotent stem cell
- NEUROG3:
-
Neurogenin 3
- NKX6.1:
-
NK6 homeobox 1
- NMD:
-
Nonsense-mediated mRNA decay
- NSG:
-
NOD-SCID-Gamma
- PDX1:
-
Pancreatic and duodenal homeobox 1
- PP:
-
Pancreatic polypeptide
- RFX6:
-
Regulatory factor X 6
- SC-islets:
-
Stem-cell-derived islets
- SLC18A1:
-
Solute carrier family 18 member A1
- SOX9:
-
SRY-box transcription factor 9
- TIRF:
-
Total internal reflection fluorescence
- VDCC:
-
Voltage-dependent calcium channel
- WT:
-
Wild-type
References
Fuchsberger C, Flannick J, Teslovich TM et al (2016) The genetic architecture of type 2 diabetes. Nature 536(7614):41–47. https://doi.org/10.1038/nature18642
Dimas AS, Lagou V, Barker A et al (2014) Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes 63(6):2158–2171. https://doi.org/10.2337/db13-0949
Bonnefond A, Unnikrishnan R, Doria A et al (2023) Monogenic diabetes. Nat Rev Dis Primer 9(1):12. https://doi.org/10.1038/s41572-023-00421-w
Aftab S, Semenec L, Chu JS-C, Chen N (2008) Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol 8(1):226. https://doi.org/10.1186/1471-2148-8-226
Sugiaman-Trapman D, Vitezic M, Jouhilahti E-M et al (2018) Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genomics 19(1):181. https://doi.org/10.1186/s12864-018-4564-6
Smith SB, Qu H-Q, Taleb N et al (2010) Rfx6 directs islet formation and insulin production in mice and humans. Nature 463(7282):775–780. https://doi.org/10.1038/nature08748
Soyer J, Flasse L, Raffelsberger W et al (2010) Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development 212(2):203–212. https://doi.org/10.1242/dev.041673
Mitchell J, Punthakee Z, Lo B et al (2004) Neonatal diabetes, with hypoplastic pancreas, intestinal atresia and gall bladder hypoplasia: search for the aetiology of a new autosomal recessive syndrome. Diabetologia 47(12):2160–2167. https://doi.org/10.1007/s00125-004-1576-3
Trott J, Alpagu Y, Tan EK et al (2020) Mitchell-Riley syndrome iPSCs exhibit reduced pancreatic endoderm differentiation due to a mutation in RFX6. Development 147(21):dev194878. https://doi.org/10.1242/dev.194878
Sansbury FH, Kirel B, Caswell R et al (2015) Biallelic RFX6 mutations can cause childhood as well as neonatal onset diabetes mellitus. Eur J Hum Genet EJHG 23(12):1744–1748. https://doi.org/10.1038/ejhg.2015.161
Skopkova M, Ciljakova M, Havlicekova Z et al (2016) Two novel RFX6 variants in siblings with Mitchell-Riley syndrome with later diabetes onset and heterotopic gastric mucosa. Eur J Med Genet 59(9):429–435. https://doi.org/10.1016/j.ejmg.2016.08.005
Piccand J, Strasser P, Hodson DJ et al (2014) Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep 9(6):2219–2232. https://doi.org/10.1016/j.celrep.2014.11.033
Chandra V, Albagli-Curiel O, Hastoy B et al (2014) RFX6 regulates insulin secretion by modulating Ca2+ homeostasis in human β cells. Cell Rep 9(6):2206–2218. https://doi.org/10.1016/j.celrep.2014.11.010
Walker JT, Saunders DC, Rai V et al (2023) Genetic risk converges on regulatory networks mediating early type 2 diabetes. Nature 624(7992):621–629. https://doi.org/10.1038/s41586-023-06693-2
Patel KA, Kettunen J, Laakso M et al (2017) Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun 8(1):888. https://doi.org/10.1038/s41467-017-00895-9
Artuso R, Provenzano A, Mazzinghi B et al (2015) Therapeutic implications of novel mutations of the RFX6 gene associated with early-onset diabetes. Pharmacogenomics J 15(1):49–54. https://doi.org/10.1038/tpj.2014.37
Mohan V, Radha V, Nguyen TT et al (2018) Comprehensive genomic analysis identifies pathogenic variants in maturity-onset diabetes of the young (MODY) patients in South India. BMC Med Genet 19(1):22. https://doi.org/10.1186/s12881-018-0528-6
Akiba K, Ushijima K, Fukami M, Hasegawa Y (2020) A heterozygous protein-truncating RFX6 variant in a family with childhood-onset, pregnancy-associated and adult-onset diabetes. Diabet Med 37(10):1772–1776. https://doi.org/10.1111/DME.13970
Imaki S, Iizuka K, Horikawa Y et al (2021) A novel RFX6 heterozygous mutation (p.R652X) in maturity-onset diabetes mellitus: a case report. J Diabetes Investig 12(10):1914–1918. https://doi.org/10.1111/JDI.13545
Tosur M, Soler-Alfonso C, Chan KM et al (2021) Exome sequencing in children with clinically suspected maturity-onset diabetes of the young. Pediatr Diabetes 22(7):960–968. https://doi.org/10.1111/pedi.13257
Elashi AA, Toor SM, Diboun I et al (2023) The genetic spectrum of Maturity-Onset Diabetes of the Young (MODY) in Qatar, a population-based study. Int J Mol Sci 24(1):130. https://doi.org/10.3390/ijms24010130
Ray D, Chatterjee N (2020) A powerful method for pleiotropic analysis under composite null hypothesis identifies novel shared loci between Type 2 Diabetes and Prostate Cancer. PLOS Genet 16(12):e1009218. https://doi.org/10.1371/journal.pgen.1009218
Kurki MI, Karjalainen J, Palta P et al (2023) FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613(7944):508–518. https://doi.org/10.1038/s41586-022-05473-8
Varshney A, Scott LJ, Welch RP et al (2017) Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc Natl Acad Sci U S A 114(9):2301–2306. https://doi.org/10.1073/pnas.1621192114
Balboa D, Iworima DG, Kieffer TJ (2021) Human pluripotent stem cells to model islet defects in diabetes. Front Endocrinol 12:149. https://doi.org/10.3389/FENDO.2021.642152/BIBTEX
Zhu Z, Li QV, Lee K et al (2016) Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell 18(6):755–768. https://doi.org/10.1016/J.STEM.2016.03.015
Nakamura T, Fujikura J, Ito R, Keidai Y, Inagaki N (2024) Human RFX6 regulates endoderm patterning at the primitive gut tube stage. PNAS Nexus 3(1):pgae001. https://doi.org/10.1093/pnasnexus/pgae001
Balboa D, Barsby T, Lithovius V et al (2022) Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat Biotechnol 40(7):1042–1055. https://doi.org/10.1038/s41587-022-01219-z
Barsby T, Ibrahim H, Lithovius V et al (2022) Differentiating functional human islet-like aggregates from pluripotent stem cells. STAR Protoc 3(4):101711. https://doi.org/10.1016/j.xpro.2022.101711
FinnGen: an expedition into genomics and medicine | FinnGen. https://www.finngen.fi/en. Accessed 14 Apr 2024
Balboa D, Saarimäki-Vire J, Borshagovski D et al (2018) Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. eLife 7:e38519. https://doi.org/10.7554/eLife.38519
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682. https://doi.org/10.1038/nmeth.2019
Stirling DR, Swain-Bowden MJ, Lucas AM, Carpenter AE, Cimini BA, Goodman A (2021) Cell Profiler 4: improvements in speed, utility and usability. BMC Bioinformatics 22(1):433. https://doi.org/10.1186/s12859-021-04344-9
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1):10–12. https://doi.org/10.14806/ej.17.1.200
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. https://doi.org/10.1093/bioinformatics/bts635
Liao Y, Smyth GK, Shi W (2019) The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47(8):e47–e47. https://doi.org/10.1093/nar/gkz114
Frankish A, Diekhans M, Jungreis I et al (2021) GENCODE 2021. Nucleic Acids Res 49(D1):D916–D923. https://doi.org/10.1093/nar/gkaa1087
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Fabregat A, Sidiropoulos K, Garapati P et al (2016) The reactome pathway knowledgebase. Nucleic Acids Res 44(D1):D481–D487. https://doi.org/10.1093/nar/gkv1351
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Schriml LM, Arze C, Nadendla S et al (2012) Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res 40(Database issue):D940–D946. https://doi.org/10.1093/nar/gkr972
Ashburner M, Ball CA, Blake JA et al (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Gu Z (2022) Complex heatmap visualization. iMeta 1(3):e43. https://doi.org/10.1002/imt2.43
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer International Publishing https://doi.org/10.1007/978-3-319-24277-4_9
Grynkiewicz G, Poenie M, Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260(6):3440–3450. https://doi.org/10.1016/S0021-9258(19)83641-4
Huopio H, Miettinen PJ, Ilonen J et al (2016) Clinical, genetic, and biochemical characteristics of early-onset diabetes in the finnish population. J Clin Endocrinol Metab 101(8):3018–3026. https://doi.org/10.1210/jc.2015-4296
Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106(25):10171–10176. https://doi.org/10.1073/pnas.0900604106
Nguyen Ba AN, Pogoutse A, Provart N, Moses AM (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10:202. https://doi.org/10.1186/1471-2105-10-202
Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R (2018) BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res 46(W1):W459–W466. https://doi.org/10.1093/nar/gky320
Qiu L, Dillman JR, Sun Q, Fei L, Abu-El-Haija M, Trout AT (2022) Pancreas volumes and predictive factors in healthy children. Pediatr Radiol 52(13):2568–2574. https://doi.org/10.1007/s00247-022-05405-8
Shih HP, Kopp JL, Sandhu M et al (2012) A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Dev Camb Engl 139(14):2488–2499. https://doi.org/10.1242/dev.078634
Zhu H, Wang G, Nguyen-Ngoc K-V et al (2023) Understanding cell fate acquisition in stem-cell-derived pancreatic islets using single-cell multiome-inferred regulomes. Dev Cell 58(9):727-743.e11. https://doi.org/10.1016/j.devcel.2023.03.011
Veres A, Faust AL, Bushnell HL et al (2019) Charting cellular identity during human in vitro β-cell differentiation. Nature 569(7756):368–373. https://doi.org/10.1038/s41586-019-1168-5
van der Meulen T, Xie R, Kelly OG, Vale WW, Sander M, Huising MO (2012) Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PloS One 7(12):e52181. https://doi.org/10.1371/journal.pone.0052181
Alhaidan Y, Christesen HT, Lundberg E, Balwi MAA, Brusgaard K (2021) CRISPR/Cas9 ADCY7 Knockout Stimulates the Insulin Secretion Pathway Leading to Excessive Insulin Secretion. Front Endocrinol 12:657873. https://doi.org/10.3389/fendo.2021.657873
Holst B, Madsen KL, Jansen AM et al (2013) PICK1 deficiency impairs secretory vesicle biogenesis and leads to growth retardation and decreased glucose tolerance. PLoS Biol 11(4):e1001542. https://doi.org/10.1371/journal.pbio.1001542
Cao M, Mao Z, Kam C et al (2013) PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol 11(4):e1001541. https://doi.org/10.1371/journal.pbio.1001541
Herlo R, Lund VK, Lycas MD et al (2018) An amphipathic helix directs cellular membrane curvature sensing and function of the BAR domain protein PICK1. Cell Rep 23(7):2056–2069. https://doi.org/10.1016/j.celrep.2018.04.074
Vanhorenbeeck V, Jenny M, Cornut J-F et al (2007) Role of the Onecut transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev Biol 305(2):685–694. https://doi.org/10.1016/j.ydbio.2007.02.027
Pedersen JK, Nelson SB, Jorgensen MC et al (2005) Endodermal expression of Nkx6 genes depends differentially on Pdx1. Dev Biol 288(2):487–501. https://doi.org/10.1016/j.ydbio.2005.10.001
Koike H, Iwasawa K, Ouchi R et al (2019) Modelling human hepato-biliary-pancreatic organogenesis from the foregut–midgut boundary. Nature 574(7776):112–116. https://doi.org/10.1038/s41586-019-1598-0
De Vas MG, Kopp JL, Heliot C, Sander M, Cereghini S, Haumaitre C (2015) Hnf1b controls pancreas morphogenesis and the generation of Ngn3+ endocrine progenitors. Dev Camb Engl 142(5):871–882. https://doi.org/10.1242/dev.110759
Piccand J, Vagne C, Blot F et al (2019) Rfx6 promotes the differentiation of peptide-secreting enteroendocrine cells while repressing genetic programs controlling serotonin production. Mol Metab 29:24–39. https://doi.org/10.1016/J.MOLMET.2019.08.007
Casellas A, Mallol C, Salavert A et al (2015) Insulin-like growth factor 2 overexpression induces β-cell dysfunction and increases beta-cell susceptibility to damage. J Biol Chem 290(27):16772–16785. https://doi.org/10.1074/jbc.M115.642041
Lee K, Kim J, Köhler M et al (2018) Blocking Ca2+ channel β3 subunit reverses diabetes. Cell Rep 24(4):922–934. https://doi.org/10.1016/j.celrep.2018.06.086
Becker A, Wardas B, Salah H et al (2021) Cavβ3 regulates Ca2+ signaling and insulin expression in pancreatic β-cells in a cell-autonomous manner. Diabetes 70(11):2532–2544. https://doi.org/10.2337/db21-0078
Yamagata K, Senokuchi T, Lu M et al (2011) Voltage-gated K+ channel KCNQ1 regulates insulin secretion in MIN6 β-cell line. Biochem Biophys Res Commun 407(3):620–625. https://doi.org/10.1016/j.bbrc.2011.03.083
Zhang J, Juhl CR, Hylten-Cavallius L et al (2020) Gain-of-function mutation in the voltage-gated potassium channel gene KCNQ1 and glucose-stimulated hypoinsulinemia - case report. BMC Endocr Disord 20(1):38. https://doi.org/10.1186/s12902-020-0513-x
Maestro MA, Cardalda C, Boj SF, Luco RF, Servitja JM, Ferrer J (2007) Distinct roles of HNF1beta, HNF1alpha, and HNF4alpha in regulating pancreas development, beta-cell function and growth. Endocr Dev 12:33–45. https://doi.org/10.1159/000109603
Plengvidhya N, Kooptiwut S, Songtawee N et al (2007) PAX4 mutations in Thais with maturity onset diabetes of the young. J Clin Endocrinol Metab 92(7):2821–2826. https://doi.org/10.1210/jc.2006-1927
Lau HH, Krentz NAJ, Abaitua F et al (2023) PAX4 loss of function increases diabetes risk by altering human pancreatic endocrine cell development. Nat Commun 14(1):6119. https://doi.org/10.1038/s41467-023-41860-z
Smith C, Moser T, Xu T, Neher E (1998) Cytosolic Ca2+ acts by two separate pathways to modulate the supply of release-competent vesicles in chromaffin cells. Neuron 20(6):1243–1253. https://doi.org/10.1016/s0896-6273(00)80504-8
Cheng C, Lu J, Cao X et al (2019) Identification of Rfx6 target genes involved in pancreas development and insulin translation by ChIP-seq. Biochem Biophys Res Commun 508(2):556–562. https://doi.org/10.1016/j.bbrc.2018.11.133
Lund A, Bagger JI, WewerAlbrechtsen NJ et al (2016) Evidence of extrapancreatic glucagon secretion in man. Diabetes 65(3):585–597. https://doi.org/10.2337/db15-1541
Funding
Open Access funding provided by University of Helsinki (including Helsinki University Central Hospital).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Acknowledgements
We thank H. Grym, A. Laitinen and R. Trokovic (Stem Cells and Metabolism Research Program, University of Helsinki) for their expert technical support. The quality control and library preparation of the RNA-seq samples were provided by the Biomedicum Functional Genomics Unit at the Helsinki Institute of Life Science and Biocenter Finland at the University of Helsinki. Novaseq sequencing was performed by the Sequencing laboratory of Institute for Molecular Medicine Finland FIMM Technology Centre, University of Helsinki. The graphical abstract was generated using BioRender. We are grateful to the family of the Mitchell–Riley syndrome patient for participating and we want to acknowledge the participants and investigators of the FinnGen study. The following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank; THL Biobank; Helsinki Biobank; Biobank Borealis of Northern Finland; Finnish Clinical Biobank Tampere; Biobank of Eastern Finland; Central Finland Biobank; Finnish Red Cross Blood Service Biobank; Terveystalo Biobank; and Arctic Biobank. All Finnish Biobanks are members of BBMRI infrastructure. Finnish Biobank Cooperative (FINBB) is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious® services managed by FINBB.
Data availability
Ultra-deep bulk RNA-seq data for pancreatic differentiation stages 3, 5 and 7 of H1 RFX6 genotypes are deposited in the Gene Expression Omnibus database with accession code GSE234289. Original western blot images are deposited at Mendeley (https://data.mendeley.com/datasets/g75drr3mgw/2). Any additional information required to reanalyse the data reported in this paper is available from the corresponding authors on request.
Funding
TO acknowledges the funding provided by the Academy of Finland (MetaStem Center of Excellence grant 312437), the Novo Nordisk Foundation, the Sigrid Juselius Foundation and the Wellcome Collaborative Award in Science. HI acknowledges the funding provided by the Doctoral Programme in Biomedicine (DPBM), Orion Research Foundation, Paulon Säätiö Foundation, Diabetestutkimussäätiö Foundation, Biomedicum Helsinki Foundation, the Finnish Cultural Foundation and Magnus Ehrnrooth Foundation. AT acknowledges project grants from the Swedish Research Council, Novo Nordisk Foundation, Diabetes Wellness Sverige, the Swedish Diabetes Foundation, Barndiabetesfonden, Family Ernfors Foundation and the strategic grant consortium Excellence of Diabetes Research in Sweden (EXODIAB). The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc., AstraZeneca UK Ltd, Biogen MA Inc., Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc., Merck Sharp & Dohme LCC, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc, Novartis AG and Boehringer Ingelheim International GmbH. The study sponsor/funders were not involved in the design of the study; the collection, analysis, and interpretation of data; writing the report; and did not impose any restrictions regarding the publication of the report.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
HI conceived and conceptualised the study, generated the H1 hESCs lines, standardised the differentiation protocols, performed the experiments, analysed data and wrote the original manuscript. DB and SE generated the iPSCs corrected cell lines and DB participated in manuscript writing. SE participated in generating the genetically edited clones, RNA purification and RT-qPCR experiments. JSV and HM performed and analysed animal experiments. MOH, OD and PEL performed and analysed cell electrophysiology experiments. JK analysed the bulk RNA-seq datasets and participated in manuscript writing. OPD and TT analysed and interpreted the FinnGen data , also participated in manuscript writing. VL conducted the morphometric analyses pipelines. TB and VC participated in analysis and manuscript writing. JU performed the insulin and C-peptide ELISA measurements. PJM is the paediatric endocrinologist in charge of the individual patient. SB and AT conceptualised the cell electrophysiology experiments and participated in the analysis and interpretation of the data and in manuscript writing. TO conceived and supervised the study, provided resources, acquired funding and participated in manuscript writing. All authors made contributions to reviewing the final version of the manuscript and approved it. HI is responsible for the integrity of the work as a whole.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ibrahim, H., Balboa, D., Saarimäki-Vire, J. et al. RFX6 haploinsufficiency predisposes to diabetes through impaired beta cell function. Diabetologia (2024). https://doi.org/10.1007/s00125-024-06163-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00125-024-06163-y