Abstract
Data generated over nearly two decades clearly demonstrate the importance of epigenetic modifications and mechanisms in the pathogenesis of type 2 diabetes. However, the role of pharmacoepigenetics in type 2 diabetes is less well established. The field of pharmacoepigenetics covers epigenetic biomarkers that predict response to therapy, therapy-induced epigenetic alterations as well as epigenetic therapies including inhibitors of epigenetic enzymes. Not all individuals with type 2 diabetes respond to glucose-lowering therapies in the same way, and there is therefore a need for clinically useful biomarkers that discriminate responders from non-responders. Blood-based epigenetic biomarkers may be useful for this purpose. There is also a need for a better understanding of whether existing glucose-lowering therapies exert their function partly through therapy-induced epigenetic alterations. Finally, epigenetic enzymes may be drug targets for type 2 diabetes. Here, I discuss whether pharmacoepigenetics is clinically relevant for type 2 diabetes based on studies addressing this topic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Research studies performed over the last two decades have identified epigenetic modifications and mechanisms that seem to play a role in the pathogenesis of type 2 diabetes [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18]. The epigenome includes DNA methylation, histone modifications and non-coding RNA [1]. There are epigenetic modifications which are stable over time and those that change due to short-term and/or long-term environmental exposures such as drugs, diet, exercise or stress, as well as ageing [19,20,21,22,23,24,25]. However, more work is needed before we fully understand the environmental regulation of the epigenome in all human cell types. Moreover, although numerous studies have investigated the role of pharmacogenetics in type 2 diabetes [26,27,28,29,30,31], the interest in pharmacoepigenetics has been limited [32]. So, what is the definition of pharmacoepigenetics and is it clinically relevant for type 2 diabetes? The meaning of pharmacoepigenetics is not set in stone but can be divided into: (1) blood-based epigenetic biomarkers that predict response or tolerance to therapy; (2) individual differences in response to therapy due to epigenetic mechanisms or variation in target cells and tissues; (3) therapies that alter the epigenome or epigenetic mechanisms, which subsequently may contribute to their effect; and (4) epigenetic therapies (Fig. 1). Below, I discuss some studies addressing these points in relation to type 2 diabetes.
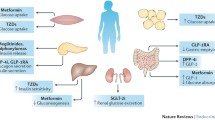
Pharmacoepigenetics in type 2 diabetes. The figure shows different aspects of pharmacoepigenetics that could be applied to type 2 diabetes prediction, response and treatment strategies. This figure is available as a downloadable slide
It is well established that all individuals do not respond to therapies in the same way. For example, ~30% of individuals with type 2 diabetes do not have a glucose-lowering response to metformin, and ~5% suffer from intolerable side effects, including gastrointestinal problems [33, 34]. Currently, there are no clinically useful biomarkers that predict response and tolerance to metformin. Nevertheless, a recent study supports the use of blood-based epigenetic biomarkers for prediction of glycaemic response and intolerance to metformin in newly diagnosed individuals with type 2 diabetes [32]. Here, increased DNA methylation of 11 CpG sites in the blood was associated with a higher risk of not responding to metformin, and increased methylation of four other CpG sites was associated with a higher risk of not tolerating metformin in drug-naive newly diagnosed individuals with type 2 diabetes. Methylation risk scores (MRS) generated based on DNA methylation levels of these 11 and four sites could clearly discriminate glycaemic responders from non-responders, and tolerant from intolerant patients to metformin therapy in three different cohorts. These results promote the further development and future use of blood-based epigenetic biomarkers for precision medicine in type 2 diabetes (Fig. 1). Therefore, pharmacoepigenetics seem to be clinically relevant for type 2 diabetes. Other factors, for example genetic variation, clinical phenotypes and gut microbiota, should further be explored, and combinations of different phenotypes may ultimately generate scores with the best predictive capacity for response to glucose-lowering therapies [26,27,28,29, 35,36,37]. Of note, in the field of cancer, both epigenetic biomarkers in blood and tissues have proven to be clinically relevant [38]. However, individual differences in response to pharmacotherapy due to epigenetic mechanisms and modifications in target tissues are, to my knowledge, not well studied in type 2 diabetes, but could be important (Fig. 1). Such epigenetic mechanisms may include DNA methylation and/or histone modifications of drug transporters, affecting the levels of these transporters in target cells and hence their ability to take up or excrete drugs.
Therapy-induced epigenetic alterations may be clinically relevant and may benefit patients (Fig. 1). Pharmacotherapies currently used for lowering blood glucose and for treatment of lipid dysregulation can alter the epigenome in tissues and cells from patients with type 2 diabetes and individuals without diabetes [25, 39,40,41,42]. For instance, individuals with type 2 diabetes who took metformin had altered DNA methylation of genes encoding the metformin transporters OCT1, OCT3 and MATE1 in the liver compared with those who did not receive any medication [39]. Short-term metformin exposure also altered DNA methylation in the blood of individuals without diabetes [25]. Additionally, incretin drugs, e.g. GLP1R agonists, prevented glucose-induced reductions in DNA methylation of NFKB1 and SOD2 in human aortic endothelial cells, which may affect vascular complications [40]. Incretin treatment was also shown to reverse epigenetic modifications associated with diabetes in rodents exposed to an impaired intrauterine environment [43]. Statin therapy was recently associated with differential DNA methylation in blood from individuals with type 2 diabetes as well as in individuals without diabetes [41, 42]. These include differential methylation of sites annotated to ABCG1, DHCR24 and SC4MOL (also known as MSMO1), which encode proteins involved in the transport and biosynthesis of cholesterol. Causal mediation analyses further suggest that DNA methylation may mediate some of statin’s effects on metabolic phenotypes [41, 42]. Overall, pharmacotherapies used for treatment of type 2 diabetes and lipid dysregulation can induce epigenetic modifications in human cells (Fig. 1). Nevertheless, further work is needed before concluding the clinical benefits or disadvantages of therapy-induced epigenetic modifications in individuals with type 2 diabetes.
Finally, can epigenetic therapies be used for treatment of type 2 diabetes (Fig. 1)? And what are epigenetic therapies? Inhibitors of epigenetic enzymes, such as DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors, fall into the category of epigenetic therapies, and such drugs are currently in use, or in clinical trials, for treatment of different cancers [44]. Interestingly, epigenetic enzymes were found to be dysregulated in cells and tissues from individuals with type 2 diabetes compared with individuals without type 2 diabetes, as well as in cells exposed to diabetogenic conditions, suggesting a potential role for epigenetic therapies also in diabetes [5, 6, 9,10,11, 45]. For example, individuals with type 2 diabetes had higher DNMT3B levels in cultured myotubes [5] and decreased TET1 expression in adipose tissue [6] vs tissue from individuals without type 2 diabetes, while palmitate exposure decreased DNMT3A and DNMT1 expression in human pancreatic islets [11]. Several studies have further shown that inhibitors of HDACs and histone demethylases, or silencing and overexpressing those enzymes, impact beta cell function and insulin secretion [9, 46,47,48,49]. For example, DNA methylation is decreased, and expression of HDAC7 increased in pancreatic islets from donors with type 2 diabetes [49]. Overexpression of Hdac7 in clonal beta cells and rat islets impaired glucose-stimulated insulin secretion, while exposure to two different HDAC inhibitors, trichostatin A (TSA) and MC1568, reversed the negative effect of Hdac7 overexpression on insulin secretion and mitochondrial function [9, 49]. MC1568 also increased glucose-stimulated insulin secretion in pancreatic islets from donors with type 2 diabetes cultured in vitro [49]. Other studies investigating the impact of the inhibition of epigenetic enzymes in muscle, adipose tissue and liver found improved metabolism and cell function [50,51,52]. However, the chronic nature of type 2 diabetes results in long-term use of therapies. It is therefore important to weigh benefits against risks, and global action of inhibitors of epigenetic enzymes may lead to intolerable side effects. More selective inhibitors and/or cell-specific delivery may represent avenues for future therapeutic purposes.
Altogether, existing literature suggests that pharmacoepigenetics may be clinically relevant for type 2 diabetes. But there is still a lot of work needed before pharmacoepigenetics in any of the research areas mentioned above may reach the clinic and help individuals with type 2 diabetes receive optimal treatment, reducing their complications and suffering.
Abbreviations
- DNMT:
-
DNA methyltransferase
- HDAC:
-
Histone deacetylase
References
Ling C, Ronn T (2019) Epigenetics in human obesity and type 2 diabetes. Cell Metab 29(5):1028–1044
Ling C, Del Guerra S, Lupi R et al (2008) Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 51(4):615–622
Ling C, Poulsen P, Simonsson S et al (2007) Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest 117(11):3427–3435
Volkov P, Bacos K, Ofori JK et al (2017) Whole-genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes 66(4):1074–1085
Davegardh C, Sall J, Benrick A et al (2021) VPS39-deficiency observed in type 2 diabetes impairs muscle stem cell differentiation via altered autophagy and epigenetics. Nat Commun 12(1):2431
Nilsson E, Jansson PA, Perfilyev A et al (2014) Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63(9):2962–2976
Nilsson E, Matte A, Perfilyev A et al (2015) Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J Clin Endocrinol Metab 100(11):E1491–E1501
Chambers JC, Loh M, Lehne B et al (2015) Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 3(7):526–534
Daneshpajooh M, Bacos K, Bysani M et al (2017) HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia 60(1):116–125
Barres R, Osler ME, Yan J et al (2009) Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 10(3):189–198
Dayeh T, Volkov P, Salo S et al (2014) Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet 10(3):e1004160
Dayeh TA, Olsson AH, Volkov P, Almgren P, Ronn T, Ling C (2013) Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia 56(5):1036–1046
Yang BT, Dayeh TA, Kirkpatrick CL et al (2011) Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 54(2):360–367
Yang BT, Dayeh TA, Volkov PA et al (2012) Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinol 26(7):1203–1212
Kirchner H, Sinha I, Gao H et al (2016) Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab 5(3):171–183
Abderrahmani A, Yengo L, Caiazzo R et al (2018) Increased hepatic PDGF-AA signaling mediates liver insulin resistance in obesity-associated type 2 diabetes. Diabetes 67(7):1310–1321
Ribel-Madsen R, Fraga MF, Jacobsen S et al (2012) Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One 7(12):e51302
Volkmar M, Dedeurwaerder S, Cunha DA et al (2012) DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J 31(6):1405–1426
Gillberg L, Perfilyev A, Brons C et al (2016) Adipose tissue transcriptomics and epigenomics in low birthweight men and controls: role of high-fat overfeeding. Diabetologia 59(4):799–812.
Ronn T, Volkov P, Davegardh C et al (2013) A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 9(6):e1003572
Katrinli S, Maihofer AX, Wani AH et al (2022) Epigenome-wide meta-analysis of PTSD symptom severity in three military cohorts implicates DNA methylation changes in genes involved in immune system and oxidative stress. Mol Psychiatry https://doi.org/10.1038/s41380-021-01398-2
Bacos K, Gillberg L, Volkov P et al (2016) Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun 7:11089
Barres R, Yan J, Egan B et al (2012) Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15(3):405–411
Feinberg AP, Irizarry RA, Fradin D et al (2010) Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med 2(49):49ra67
Elbere I, Silamikelis I, Ustinova M et al (2018) Significantly altered peripheral blood cell DNA methylation profile as a result of immediate effect of metformin use in healthy individuals. Clin Epigenetics 10(1):156
Zhou K, Yee SW, Seiser EL et al (2016) Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet 48(9):1055–1059
Zhou K, Donnelly L, Yang J et al (2014) Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol 2(6):481–487
Rotroff DM, Yee SW, Zhou K et al (2018) Genetic variants in CPA6 and PRPF31 are associated with variation in response to metformin in individuals with type 2 diabetes. Diabetes. 67(7):1428–1440
Jablonski KA, McAteer JB, de Bakker PI et al (2010) Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 59(10):2672–2681
GoDarts, Group UDPS, Wellcome Trust Case Control C, Zhou K, Bellenguez C, Spencer CC et al (2011) Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 43(2):117–120
Dujic T, Zhou K, Tavendale R, Palmer CN, Pearson ER (2016) Effect of serotonin transporter 5-HTTLPR polymorphism on gastrointestinal intolerance to metformin: a GoDARTS study. Diabetes Care 39(11):1896–1901
Garcia-Calzon S, Perfilyev A, Martinell M et al (2020) Epigenetic markers associated with metformin response and intolerance in drug-naive patients with type 2 diabetes. Sci Transl Med 12:561
Cook MN, Girman CJ, Stein PP, Alexander CM (2007) Initial monotherapy with either metformin or sulphonylureas often fails to achieve or maintain current glycaemic goals in patients with type 2 diabetes in UK primary care. Diabet Med 24(4):350–358
Kahn SE, Haffner SM, Heise MA et al (2006) Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N Engl J Med 355(23):2427–2443
Donnelly LA, Doney AS, Hattersley AT, Morris AD, Pearson ER (2006) The effect of obesity on glycaemic response to metformin or sulphonylureas in type 2 diabetes. Diabet Med 23(2):128–133
Dujic T, Zhou K, Donnelly LA, Tavendale R, Palmer CN, Pearson ER (2015) Association of Organic Cation Transporter 1 with intolerance to metformin in type 2 diabetes: a GoDARTS study. Diabetes 64(5):1786–1793
Dujic T, Causevic A, Bego T et al (2016) Organic cation transporter 1 variants and gastrointestinal side effects of metformin in patients with type 2 diabetes. Diabet Med 33(4):511–514
Oliver J, Garcia-Aranda M, Chaves P et al (2021) Emerging noninvasive methylation biomarkers of cancer prognosis and drug response prediction. Semin Cancer Biol https://doi.org/10.1016/j.semcancer.2021.03.012
Garcia-Calzon S, Perfilyev A, Mannisto V et al (2017) Diabetes medication associates with DNA methylation of metformin transporter genes in the human liver. Clin Epigenetics 9:102
Scisciola L, Rizzo MR, Cataldo V et al (2020) Incretin drugs effect on epigenetic machinery: new potential therapeutic implications in preventing vascular diabetic complications. FASEB J 34(12):16489–16503
Schrader S, Perfilyev A, Martinell M, Garcia-Calzon S, Ling C (2021) Statin therapy is associated with epigenetic modifications in individuals with type 2 diabetes. Epigenomics. 13(12):919–925
Ochoa-Rosales C, Portilla-Fernandez E, Nano J et al (2020) Epigenetic link between statin therapy and type 2 diabetes. Diabetes Care 43(4):875–884
Pinney SE, Jaeckle Santos LJ, Han Y, Stoffers DA, Simmons RA (2011) Exendin-4 increases histone acetylase activity and reverses epigenetic modifications that silence Pdx1 in the intrauterine growth retarded rat. Diabetologia 54(10):2606–2614
Cheng Y, He C, Wang M et al (2019) Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 4(1):62
Nilsson E, Vavakova M, Perfilyev A et al (2021) Differential DNA Methylation and Expression of MicroRNAs in Adipose Tissue from Twin Pairs Discordant for Type 2 Diabetes. Diabetes 70(10):2402–2418
Backe MB, Andersson JL, Bacos K et al (2018) Lysine demethylase inhibition protects pancreatic beta cells from apoptosis and improves beta-cell function. Mol Cell Endocrinol 460:47–56
Backe MB, Jin C, Andreone L et al (2019) The lysine demethylase KDM5B regulates islet function and glucose homeostasis. J Diabetes Res 2019:5451038
Lundh M, Galbo T, Poulsen SS, Mandrup-Poulsen T (2015) Histone deacetylase 3 inhibition improves glycaemia and insulin secretion in obese diabetic rats. Diabetes Obes Metab 17(7):703–707
Daneshpajooh M, Eliasson L, Bacos K, Ling C (2018) MC1568 improves insulin secretion in islets from type 2 diabetes patients and rescues beta-cell dysfunction caused by Hdac7 upregulation. Acta Diabetol 55(12):1231–1235
Galmozzi A, Mitro N, Ferrari A et al (2013) Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes 62(3):732–742
Lee SJ, Choi SE, Lee HB et al (2020) A class I histone deacetylase inhibitor attenuates insulin resistance and inflammation in palmitate-treated C2C12 Myotubes and muscle of HF/HFr diet mice. Front Pharmacol 11:601448
Gao Z, Yin J, Zhang J et al (2009) Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58(7):1509–1517
Acknowledgements
I want to thank Dr. Tina Rönn, Department of Clinical Sciences, Lund University Diabetes Centre, Lund University, Scania University Hospital, 205 02 Malmö, Sweden, for helpful comments on this opinion piece.
Authors’ relationships and activities
The author declares that there are no relationships or activities that might bias, or be perceived to bias, their work.
Contribution statement
CL is the sole contributor.
Funding
Open access funding provided by Lund University. Work in the author’s laboratory is supported by grants from the EFSD, the Novo Nordisk foundation, The Swedish Research Council, Region Skåne (ALF), ERC-Co Grant (PAINTBOX, No 725840), Exodiab, Swedish Foundation for Strategic Research for IRC15-0067 and the Swedish Diabetes Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PPTX 350 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ling, C. Pharmacoepigenetics in type 2 diabetes: is it clinically relevant?. Diabetologia 65, 1849–1853 (2022). https://doi.org/10.1007/s00125-022-05681-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-022-05681-x