Abstract
The current classification of diabetes, based on hyperglycaemia, islet-directed antibodies and some insufficiently defined clinical features, does not reflect differences in aetiological mechanisms and in the clinical course of people with diabetes. This review discusses evidence from recent studies addressing the complexity of diabetes by proposing novel subgroups (subtypes) of diabetes. The most widely replicated and validated approach identified, in addition to severe autoimmune diabetes, four subgroups designated severe insulin-deficient diabetes, severe insulin-resistant diabetes, mild obesity-related diabetes and mild age-related diabetes subgroups. These subgroups display distinct patterns of clinical features, disease progression and onset of comorbidities and complications, with severe insulin-resistant diabetes showing the highest risk for cardiovascular, kidney and fatty liver diseases. While it has been suggested that people in these subgroups would benefit from stratified treatments, RCTs are required to assess the clinical utility of any reclassification effort. Several methodological and practical issues also need further study: the statistical approach used to define subgroups and derive recommendations for diabetes care; the stability of subgroups over time; the optimal dataset (e.g. phenotypic vs genotypic) for reclassification; the transethnic generalisability of findings; and the applicability in clinical routine care. Despite these open questions, the concept of a new classification of diabetes has already allowed researchers to gain more insight into the colourful picture of diabetes and has stimulated progress in this field so that precision diabetology may become reality in the future.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.

Rationale for diabetes reclassification
The observation that people with diabetes have different phenotypes has led to repeated attempts to classify the main diabetes types [1, 2]. Electronic supplementary material (ESM) Table 1 summarises the key efforts in this evolution. People with diabetes show a broad variation in the main features of diabetes (i.e. insulin resistance and beta cell dysfunction [3,4,5,6]) as a result of the combined effects of (epi)genetic, environmental and lifestyle factors and their different contributions in different individuals. A recently proposed ‘palette model’ conceptualises the interaction of these factors [3]. People at risk of diabetes may have impairments in multiple processes such as islet development, islet function, autoimmunity, inflammation, insulin sensitivity, incretin activity and adipose tissue function (considered as ‘base colours’). Every individual is positioned somewhere within the spectrum of the phenotypic variation of each trait as determined by their genetic variation and non-genetic exposures modifying these processes, and the sum (or mixture) of all trait variations represents the overall estimate of metabolic health and diabetes status [3]. However, this pathophysiological heterogeneity is not captured by current position statements and guidelines for diagnosis and treatment of diabetes [7, 8].
Differences in risk factors and pathophysiological mechanisms are thought to drive the heterogeneity in preclinical abnormalities, prevalence of comorbidities and clinical complications already seen at diagnosis of diabetes [5, 9]. People with diabetes further vary in the progression of their disease and in the incidence of diabetes-related complications despite comparable glycaemic control.
Any reclassification effort should be seen as an example of precision medicine or ‘precision diabetology’ aiming to deconstruct the heterogeneity of diabetes. Advances in the management of monogenic forms of diabetes (neonatal diabetes, MODY) represent a successful proof-of-concept for a reclassification of diabetes [10]. Currently, however, relatively few people with diabetes are affected by monogenic diabetes so this may serve as an example for personalised medicine based on mutations in single genes [11] rather than the precision medicine approach that is required for type 1 and type 2 diabetes, which are both polygenic and multifactorial [12].
In the context of type 1 and type 2 diabetes, the ultimate purpose of precision diabetology is the development of stratified prevention and treatment for subgroups of people with different risk profiles. These options range from refined screening and monitoring intervals, recommendations for tailored lifestyle interventions, to targeted but not individualised drug treatment. Clinical benefits envisaged include fewer adverse effects and ideally a delay of the onset of diabetes and its complications, lower morbidity and mortality and an economic use of resources [13].
The aim of this review is to provide an up-to-date, concise overview of studies on diabetes reclassification, their implications, and also inherent practical and methodological challenges, with a specific focus on recent definitions of subgroups of type 2 diabetes and the risk of complications in these subgroups. With respect to subgroups and endotypes of type 1 diabetes, we would like to refer the reader to recent excellent reviews covering aspects of precision diabetology for this diabetes type [8, 14].
Variability of disease presentation and progression
One approach to study the heterogeneity of diabetes relies on cohorts of people included at or shortly after the diagnosis of diabetes [5]. Even though the duration of hyperglycaemia before diabetes diagnosis is unknown, these cohorts allow the investigation of clinical characteristics that are not yet confounded by long-term excessive hyperglycaemia and pharmacological treatment. Examples are the German Diabetes Study (GDS [5]), the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS [15]) and the All New Diabetics in Scania (ANDIS [6]) cohorts.
The comprehensive phenotyping in the GDS, using gold-standard methodology, demonstrated large interindividual differences in people within 12 months of their diagnosis of diabetes regarding insulin sensitivity, beta cell function, islet-directed autoantibodies, blood lipids and BP [5, 16]. The variability in disease severity or progression is reflected by differences between subgroups of type 2 diabetes regarding diabetes-related complications such as chronic kidney disease (CKD), distal sensorimotor polyneuropathy (DSPN), cardiovascular autonomic neuropathy (CAN), retinopathy and non-alcoholic fatty liver disease (NAFLD) [5, 16, 17].
The VNDS enrols people with type 2 diabetes within 6 months of their diagnosis. Assessment of diabetes-related complications indicated a high variability in their presentation and progression [15, 18]; the prevalence of one or more diabetes-related complications (CVD, nephropathy, DSPN, CAN, retinopathy) already present at study enrolment was found to be 49.2% [9].
The ANDIS cohort includes incident cases of diabetes and reported a marked prevalence of NAFLD at baseline (although only based on surrogate measurement by alanine aminotransferase) and the development of CKD, retinopathy and CVD in the first years after the diagnosis of diabetes [6].
In addition, people with diabetes differ with respect to inherited factors. The application of a polygenic risk score (PRS) based on >136,000 variants in the UK Biobank demonstrated a prevalence of type 2 diabetes of 1.2% and 11.2% in the lowest and highest 2.5% of the PRS distribution, respectively, revealing an almost tenfold difference [19].
Taken together, these studies highlight the substantial variability in the pathogenic and clinical characteristics of the large population of people commonly designated as having type 2 diabetes.
Novel subtypes of diabetes reflecting differences in disease development and progression
Reclassification methods
From a methodological perspective, different clustering algorithms have been used to reclassify people with diabetes [20,21,22,23,24,25]. Topology-based analysis [21] and Bayesian non-negative matrix factorisation clustering [22] are widely applied procedures for discovering groups of related observations (e.g. subgroups of people with diabetes) using high-dimensional data such as electronic medical records or omics data. Cluster analysis based on the k-means or the partitioning around medoids methods [6, 23] break large datasets up into subgroups by minimising the distance between data points labelled to be in a cluster and a point designated as the centre of that cluster. These methods require that the optimal number of clusters (k) needs to be known a priori (i.e. evaluated with other methods). In contrast, latent-class trajectory analysis is a longitudinal analysis method using repeated measures of dependent variables as a function of time to identify subgroups of people who differ in trajectories (e.g. in glucose response curves) [24, 25].
In addition to these methods, reclassification studies made use of datasets that differed widely in the type and number of variables. One clustering approach, using high-dimensional electronic medical records and extensive genotype data, identified three subtypes of type 2 diabetes enriched in CVD, nephropathy, retinopathy, neurological diseases and cancer [21]. A second approach used data for 94 type 2 diabetes-associated gene variants and 47 diabetes-related traits to subgroup genetic loci according to mechanistic pathways and to relate the clinical characteristics of people with type 2 diabetes to their genetic risk scores [22]. This study found two clusters of genetic loci related to insulin deficiency and three related to insulin resistance. Individuals with high genetic risk scores in the respective clusters also differed in obesity, lipids, hypertension, kidney function and CVD [22].
From a clinical perspective, clustering algorithms based on available patient data would be highly attractive. One study used latent-class trajectory analysis based on mixed-meal tolerance tests in people with newly diagnosed type 2 diabetes [25] and identified three subgroups based on their glucose response patterns. Thus, this method represents another classification approach closely related to insulin resistance and insulin secretion as the pathophysiological hallmarks of type 2 diabetes.
Diabetes subgroups
The most frequently replicated study in this field used both hierarchical and k-means clustering in Swedish people with newly diagnosed diabetes, with the following six variables as input: GAD antibodies; age at diagnosis; BMI at diagnosis; HbA1c; and HOMA-2 estimates of insulin resistance and beta cell function calculated from fasting glucose and C-peptide [6]. The resulting subgroups (subtypes) were designated as severe autoimmune diabetes (SAID), severe insulin-deficient diabetes (SIDD), severe insulin-resistant diabetes (SIRD), mild obesity-related diabetes (MOD) and mild age-related diabetes (MARD) [6] (Table 1). This concept has been replicated in cohorts from Europe, North America and Asia despite varying disease duration since diabetes diagnosis [26,27,28,29,30,31,32]. The SAID subgroup comprises people who are otherwise classified as having type 1 diabetes (including those previously termed latent autoimmune diabetes of adults), whereas SIDD, SIRD, MOD and MARD represent novel entities of type 2 diabetes. The subgroups also differ with respect to prevalence and/or risk of complications (Table 1). In line with the predominant insulin deficiency, ketoacidosis at diagnosis is most frequent in SAID and SIDD [6]. Retinopathy, DSPN and CAN are observed most often in SIDD [6, 26], while CKD and NAFLD are most prevalent in SIRD [6, 26, 29]. Adjusted risk ratios for prevalent erectile dysfunction are highest for SIDD and SIRD [33]. Although the subgroups differ in their cardiovascular risk, these differences did not remain statistically significant after adjustment for age and sex in the ANDIS cohort [6] or after more comprehensive adjustment for multiple covariables in a Japanese cohort [29].
Subgroup variables
The clinical relevance of the novel subgroups has been assessed in multiple cohorts including ethnically diverse populations that lack some of the aforementioned clustering variables (most often C-peptide measurements). Partial replication of the subgroup classification and differential risk of complications was reported in cohorts from Europe [34, 35], the USA [36, 37], Mexico [38], Latin America and the Caribbean [39], India [40] and China [41] and in large international trial populations (DEVOTE/LEADER/SUSTAIN-6 [42]) (Table 2). At present it is not known whether C-peptide or insulin are required to identify SIRD, so it would be informative to compare different combinations of clustering variables (e.g. with and without C-peptide or insulin) in the same cohorts, to better understand their relevance for reclassification.
Only a few studies have explored biomarkers and pathways underlying differences between subgroups that could determine susceptibility to diabetes-related complications. Given the role of lipid metabolism in diabetes, it is noteworthy that serum triacylglycerol levels were found to be highest and HDL-cholesterol levels lowest in SIRD, while there were no differences in total or LDL-cholesterol [26, 29, 43]. Circulating levels of angiopoietin-like protein 8 (ANGPTL8), a regulator of lipid metabolism, were higher in SIDD, SIRD and MARD than in MOD [44]. However, these differences were not adjusted for the clustering variables.
Subgroup differences in inflammation
Circulating levels of high-sensitivity C-reactive protein (hsCRP) were highest in SIRD and MOD [29]. The association of circulating triacylglycerols and inflammatory processes with insulin resistance is in line with the uniform mechanism underlying common insulin resistance in humans [45]. A multimarker approach in the GDS found that 23 biomarkers of inflammation differed between the subgroups, with biomarker levels in general being highest in SIRD and lowest in SIDD [46]. After adjustment for the clustering variables, serum caspase-8 (CASP-8), S100 calcium-binding protein A12 (EN-RAGE) and IL-6 showed at least one pairwise difference between the subgroups. The association between inflammation and insulin resistance reflects the contribution of inflammation-related processes to SIRD, whereas inflammatory processes appear less relevant in SIDD [46]. A second study in this cohort showed that the SIRD subgroup also had high leucocyte numbers and the highest CD4+ T cell percentages, thereby demonstrating different immune cell frequencies between subgroups and highlighting the proinflammatory characteristics of SIRD [47]. Of note, studies on autoimmune diabetes identified both genetic and epigenetic determinants of T cell function, with effects on gene expression [48, 49]. (Epi)genetic variation and its impact on transcriptomes in immune cells will require more detailed analyses with respect to relevance in disease aetiology in the other subgroups.
Genetic predisposition
There is evidence that the subgroups may differ in their associations with gene variants predisposing to diabetes. The HLA SNP rs2854275 showed the same association with SAID as with type 1 diabetes in previous studies, but not with SIDD, pointing towards different aetiologies of insulin deficiency in the subgroups [6]. Both the TCF7L2 SNP rs7903146, which is known for its association with type 2 diabetes, and a genetic score for type 2 diabetes were associated with SIDD, MOD and MARD but not with SIRD [6]. A genetic risk score for insulin secretion was associated with MOD and MARD (and nominally with SIDD) but again not with SIRD [6]. This suggests a more pronounced role for genetic predisposition to SIDD, MOD and MARD and a stronger role for environmental determinants in SIRD. Of note, individuals with SIRD were more frequently carriers of the G allele of rs738409 in PNPLA3, the gene encoding patatin-like phospholipase domain-containing-3, which is characterised by its positive association with hepatic fat content [43] and may contribute to the relationship between SIRD and progression of NAFLD to fibrosis [6, 26, 29]. Thus, genetic analyses corroborate the difference between SAID and the other subgroups but suggest unique mechanisms that might distinguish SIRD from SIDD, MOD and MARD.
At present, the only study integrating genetic, metabolomic, lipidomic and proteomic data to compare diabetes subtypes was based on different clustering variables (age, BMI, HbA1c, HDL-cholesterol, and random or fasting C-peptide). SIRD showed the most distinct molecular signature, mostly related to insulin resistance, lipids and inflammation [50].
Subgroups of individuals with different risk of progression to type 2 diabetes and complications
Trajectory analyses show that changes in metabolic and inflammation-related biomarkers start >10 years before the onset of type 2 diabetes [51,52,53]. Therefore, it is also of high clinical relevance to identify subgroups of individuals at different risk for diabetes and for complications, which may start even before the manifestation of diabetes. Latent-class trajectory analysis using OGTTs in people without diabetes revealed four subgroups that differed in anthropometric, metabolic and inflammation-related variables [24], but this study did not analyse diabetes-related complications.
A recent study in a cohort of individuals at elevated risk of type 2 diabetes explored the pathophysiological heterogeneity before clinical diabetes onset [23]. Participants from the Tübingen Family Study (TUEF) and Tübingen Lifestyle Intervention Program (TULIP) underwent clustering based on OGTT, MRI (body fat distribution, liver fat), serum lipids and a PRS for type 2 diabetes. This study found six subphenotypes differing in diabetes-related variables: 1, low risk; 2, very low risk; 3, beta cell failure; 4, low-risk obese; 5, high-risk insulin-resistant fatty liver; 6, high-risk visceral fat nephropathy [23]. Results were replicated in the Whitehall II cohort using a reduced set of clustering variables. Overall, clusters 3, 5 and 6 showed higher glucose levels at baseline but only clusters 3 and 5 had an increased incidence of type 2 diabetes. Clusters 3, 5 and 6 featured the highest CKD risk and higher intima–media thickness, and clusters 5 and 6 had the highest all-cause mortality. Data from Whitehall II indicated that individuals from the low-risk clusters 1, 2 and 4 transitioned to MOD and MARD with diabetes onset, whereas individuals from the high-risk cluster 6 transitioned to SIRD [23]. Thus, clustering approaches can also identify subphenotypes with respect to glycaemic, renal, cardiovascular and all-cause mortality risk, corresponding to previous findings for overt diabetes [6, 26].
Translation into clinical practice: therapeutic implications
RCTs are required to evaluate the clinical relevance of reclassification efforts. Until data from subgroup-specific RCTs are available, it is only possible to investigate in cohort studies [6, 29] or intervention trials, such as A Diabetes Outcome Progressive Trial (ADOPT) [28], whether individuals allocated to subgroups differ in their treatment at study baseline or their treatment responses, respectively (Table 3). Importantly, the high frequency of individuals without initial glucose-lowering treatment, the low frequency of insulin use and the shortest time to reach the HbA1c target were similar for SIRD, MOD and MARD and correspond to their less pronounced insulin deficiency compared with SAID and SIDD (Table 3). Glycaemic deterioration may thus suggest a milder progression of disease for SIRD, MOD and MARD. However, the higher risk for several complications in SIRD clearly indicates the need for treatment intensification addressing CKD, CVD and NAFLD (e.g. by sodium–glucose cotransporter 2 inhibitors [SGLT2is] and glucagon-like peptide-1 receptor agonists [GLP-1RAs]), as well as targeting insulin resistance (e.g. by future insulin sensitisers) (Fig. 1). Given the proinflammatory profile and the high risk of complications in SIRD, novel therapies targeting inflammatory pathways, as developed for people at high cardiovascular risk [54, 55], could also be considered in the future. Initially, lifestyle modification and metformin are sufficient for treating MOD and MARD. Nevertheless, MOD may specifically benefit from weight loss intervention by hypo-energetic diets and drugs, whereas MARD may be better treated by nutrition avoiding further ageing-related sarcopenia (Fig. 1).
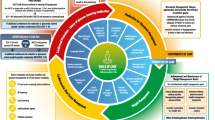
Possible future implications of precision diabetology based on the novel diabetes subgroups. Although the utility of the concept needs to be evaluated in RCTs, one may speculate on the potential implications of a new (sub)classification of diabetes for tailored diagnosis, prevention and treatment. Individuals in the different diabetes subgroups differ in their susceptibility to developing specific complications. The different (pathophysiological) phenotypes may also differ in their response to lifestyle-related and pharmacological strategies. SAID requires early introduction of insulin supplementation, whereas SIDD may also benefit from a dipeptidyl peptidase 4 inhibitor (DPP4i) or, when cost is a major issue, a sulfonylurea. SIRD and MOD would benefit from medication that induces weight loss (SGLT2i, GLP-1RA, dual agonist) or also addresses risk of CVD or nephropathy (SGLT2i, GLP-1RA). Providing that safety and efficacy have been established, new insulin sensitisers (e.g. peroxisome proliferator activator receptor agonists) or anti-inflammatory drugs could also improve targeted treatment of SIRD. On the other hand, individuals with MARD should receive treatments avoiding weight loss and sarcopenia (e.g. protein-balanced diets and moderate resistance training). PPARa, peroxisome proliferator activator receptor agonist. This figure is available as a downloadable slide
Currently, it is not clear whether our knowledge on mechanisms (and adverse effects) of these drugs will translate into subgroup-specific treatment benefits [56]. However, the large number of ongoing RCTs using novel therapeutic agents targeting insulin secretion, insulin resistance, liver metabolism and other mechanisms that differ between the subgroups holds promise for precision healthcare [57].
So far, only the Look AHEAD (Action for Health in Diabetes) study analysed the differential response to lifestyle intervention [37]. Individuals with type 2 diabetes were allocated to four subgroups, which are not directly comparable with the previously described subgroups [6, 26] due to differences in clustering variables. Randomisation to intensive lifestyle intervention was associated with increased cardiovascular risk in the subgroup characterised by the poorest glucose control and most frequent use of insulin [37]. Thus, subgroups may differ in their response to non-pharmacological treatment, emphasising the differential need for pharmacological treatment intensification to prevent diabetes-related complications.
Methodological aspects and open questions
The text box above gives an overview of key gaps in our current knowledge, resulting open questions, and future directions in this field; some of the methodological aspects are also briefly discussed here. Any effort made concerning reclassification into subgroups has a strong conceptual appeal because it is easy to communicate and implement once RCTs have shown subgroup-specific differences to non-pharmacological and pharmacological interventions. However, this approach assumes a certain degree of homogeneity within, and clear differences between, subgroups, whereas in reality the characteristics of individuals from different clusters partially overlap [3]. The subgroup approach is also limited by the fact that subgroup assignment requires the availability of all clustering variables. Finally, the utility of subgroups depends on their stability. An analysis in the GDS demonstrated that 23% of the study participants migrated into a different subgroup within the first 5 years after the diagnosis of diabetes [26].
An alternative strategy in precision diabetology may be based on statistical models using continuous risk factors [28, 58, 68]. In a secondary analysis of RCTs [28], age at diabetes diagnosis and renal function at baseline were better predictors of disease progression than the subgroup assignment according to Ahlqvist et al [6]. Thus, specific phenotypic measures to predict glycaemic progression, onset of complications and treatment response could be used to optimise diabetes care in an individualised approach. The risk assessment could be updated regularly to take into account disease progression, with corresponding treatment changes. However, these models would only be useful for optimising one specific outcome such as glycaemic progression or the development of a predefined complication unless they were a priori designed to predict a composite endpoint comprising different outcomes based on the patients’ preferences. Currently, such an approach remains challenging because it requires a huge amount of individual-participant data to develop the underlying models.
One general criticism of the aforementioned reclassification strategies refers to their use of phenotypic data that depend on disease progression, lifestyle and medication and therefore necessitate regular adaptation. In contrast, genotypic data are stable over time and are more likely to be related to causal mechanisms [22]. However, the proportion of diabetes risk that can be explained by environmental risk factors is still greater than the proportion that can be attributed to known genetic risk variants. This means that people with large differences in genetic risk scores show minor phenotypic differences that can be overcome by modifying exogenous risk factors [69, 70]. Currently, it is unknown which of the two approaches or alternatively a combination of both phenotypic and genotypic reclassification would provide the best benefit.
Irrespective of all methodological and practical issues, it is important to emphasise the following points: (1) clinical decision making is always binary at the end (i.e. resulting in the decision to treat or not to treat and in the selection of certain non-pharmacological or pharmacological interventions) and (2) any approach to reclassify diabetes must result in diabetes prevention and care superior to that received under the established classification. The ongoing initiative of the ADA and the EASD on precision medicine in diabetes will provide a detailed roadmap for future studies and application of tailored diagnostics, prevention and treatment on the road to precision diabetology [13].
Conclusions
The heterogeneity of diabetes, particularly type 2 diabetes, is evident from differences in multiple pathophysiological and clinical features. Recent studies provided novel insights into interindividual differences by clustering people with diabetes into five subgroups, which are reproducible and associated with different trajectories in disease progression and onset of diabetes-related complications including CKD, retinopathy, CVD, NAFLD and neuropathies. Based on the current evidence, it is possible to propose innovative stratified prevention and treatment approaches at least for some of these subgroups (Fig. 1). However, the ultimate test of the utility of precision diabetology will require RCTs to demonstrate whether the probability-based assignment to subgroups and subsequent subgroup-specific prevention or treatment is indeed superior to that proposed by the current guidelines. Furthermore, future studies should address methodological issues, in particular on the best precision diabetology approaches, and also uncertainties regarding the transethnic generalisability of the current findings.
Abbreviations
- ADOPT:
-
A Diabetes Outcome Progressive Trial
- AHEAD:
-
Action for Health in Diabetes
- ANDIS:
-
All New Diabetics in Scania
- ANGPTL8:
-
Angiopoietin-like protein 8
- CAN:
-
Cardiovascular autonomic neuropathy
- CASP-8:
-
Caspase-8
- CKD:
-
Chronic kidney disease
- DSPN:
-
Distal sensorimotor polyneuropathy
- EN-RAGE:
-
S100 calcium-binding protein A12
- GDS:
-
German Diabetes Study
- GLP-1RA:
-
Glucagon-like peptide-1 receptor agonist
- hsCRP:
-
High-sensitivity C-reactive protein
- MARD:
-
Mild age-related diabetes
- MOD:
-
Mild obesity-related diabetes
- NAFLD:
-
Non-alcoholic fatty liver disease
- PRS:
-
Polygenic risk score
- SAID:
-
Severe autoimmune diabetes
- SGLT2i:
-
Sodium–glucose cotransporter 2 inhibitor
- SIDD:
-
Severe insulin-deficient diabetes
- SIRD:
-
Severe insulin-resistant diabetes
- VNDS:
-
Verona Newly Diagnosed Type 2 Diabetes Study
References
Gale EA (2001) The discovery of type 1 diabetes. Diabetes 50(2):217–226. https://doi.org/10.2337/diabetes.50.2.217
Gale EA (2013) Is type 2 diabetes a category error? Lancet 381(9881):1956–1957. https://doi.org/10.1016/S0140-6736(12)62207-7
McCarthy MI (2017) Painting a new picture of personalised medicine for diabetes. Diabetologia 60(5):793–799. https://doi.org/10.1007/s00125-017-4210-x
Pearson ER (2019) Type 2 diabetes: a multifaceted disease. Diabetologia 62(7):1107–1112. https://doi.org/10.1007/s00125-019-4909-y
Szendroedi J, Saxena A, Weber KS et al (2016) Cohort profile: the German diabetes study (GDS). Cardiovasc Diabetol 15:59. https://doi.org/10.1186/s12933-016-0374-9
Ahlqvist E, Storm P, Käräjämäki A et al (2018) Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6(5):361–369. https://doi.org/10.1016/S2213-8587(18)30051-2
Davies MJ, D’Alessio DA, Fradkin J et al (2018) Management of hyperglycaemia in type 2 diabetes, 2018. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of diabetes (EASD). Diabetologia 61(12):2461–2498. https://doi.org/10.1007/s00125-018-4729-5 Erratum in: Diabetologia 2019;62(5):873
Redondo MJ, Hagopian WA, Oram R et al (2020) The clinical consequences of heterogeneity within and between different diabetes types. Diabetologia 63(10):2040–2048. https://doi.org/10.1007/s00125-020-05211-7
Bonora E, Trombetta M, Dauriz M et al (2020) Chronic complications in patients with newly diagnosed type 2 diabetes: prevalence and related metabolic and clinical features: the Verona newly diagnosed type 2 diabetes study (VNDS) 9. BMJ Open Diabetes Res Care 8(1):e001549. https://doi.org/10.1136/bmjdrc-2020-001549
Prasad RB, Groop L (2019) Precision medicine in type 2 diabetes. J Intern Med 285(1):40–48. https://doi.org/10.1111/joim.12859
Gloyn AL, Drucker DJ (2018) Precision medicine in the management of type 2 diabetes. Lancet Diabetes Endocrinol 6(11):891–900. https://doi.org/10.1016/S2213-8587(18)30052-4
Barroso I, McCarthy MI (2019) The genetic basis of metabolic disease. Cell 177(1):146–161. https://doi.org/10.1016/j.cell.2019.02.024
Chung WK, Erion K, Florez JC et al (2020) Precision medicine in diabetes: a consensus report from the American Diabetes Association (ADA) and the European Association for the Study of diabetes (EASD). Diabetologia 63(9):1671–1693. https://doi.org/10.1007/s00125-020-05181-w
Battaglia M, Ahmed S, Anderson MS et al (2020) Introducing the Endotype concept to address the challenge of disease heterogeneity in type 1 diabetes. Diabetes Care 43(1):5–12. https://doi.org/10.2337/dc19-0880
Zoppini G, Cacciatori V, Raimondo D et al (2015) Prevalence of cardiovascular autonomic neuropathy in a cohort of patients with newly diagnosed type 2 diabetes: the Verona newly diagnosed type 2 diabetes study (VNDS). Diabetes Care 38(8):1487–1493. https://doi.org/10.2337/dc15-0081
Zaharia OP, Bobrov P, Strassburger K et al (2018) Metabolic characteristics of recently diagnosed adult-onset autoimmune diabetes mellitus. J Clin Endocrinol Metab 103(2):429–437. https://doi.org/10.1210/jc.2017-01706
Maalmi H, Herder C, Strassburger K et al (2020) Biomarkers of inflammation and glomerular filtration rate in individuals with recent-onset type 1 and type 2 diabetes. J Clin Endocrinol Metab 105(12):e4370–e4381. https://doi.org/10.1210/clinem/dgaa622
Zoppini G, Trombetta M, Pastore I et al (2021) Glomerular filtration rate decline in T2DM following diagnosis. The Verona newly diagnosed diabetes Study-12. Diabetes Res Clin Pract 22:108778. https://doi.org/10.1016/j.diabres.2021.108778
Mahajan A, Taliun D, Thurner M et al (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50(11):1505–1513. https://doi.org/10.1038/s41588-018-0241-6
Sarría-Santamera A, Orazumbekova B, Maulenkul T, Gaipov A, Atageldiyeva K (2020) The identification of diabetes mellitus subtypes applying cluster analysis techniques: a systematic review. Int J Environ Res Public Health 17(24):9523. https://doi.org/10.3390/ijerph17249523
Li L, Cheng WY, Glicksberg BS et al (2015) Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 7(311):311ra174. https://doi.org/10.1126/scitranslmed.aaa9364
Udler MS, Kim J, von Grotthuss M et al (2018) Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med 15(9):e1002654. https://doi.org/10.1371/journal.pmed.1002654
Wagner R, Heni M, Tabák AG et al (2021) Pathophysiology-based subphenotyping of individuals at elevated risk for type 2 diabetes. Nat Med 27(1):49–57. https://doi.org/10.1038/s41591-020-1116-9
Hulman A, Witte DR, Vistisen D et al (2018) Pathophysiological characteristics underlying different glucose response curves: a latent class trajectory analysis from the prospective EGIR-RISC study. Diabetes Care 41(8):1740–1748. https://doi.org/10.2337/dc18-0279
Obura M, Beulens JWJ, Slieker R et al (2020) Post-load glucose subgroups and associated metabolic traits in individuals with type 2 diabetes: an IMI-DIRECT study. PLoS One 15(11):e0242360. https://doi.org/10.1371/journal.pone.0242360
Zaharia OP, Strassburger K, Strom A et al (2019) Risk of diabetes-associated diseases in subgroups of patients with recent-onset diabetes: a 5-year follow-up study. Lancet Diabetes Endocrinol 7(9):684–694. https://doi.org/10.1016/S2213-8587(19)30187-1
Zou X, Zhou X, Zhu Z, Ji L (2019) Novel subgroups of patients with adult-onset diabetes in Chinese and US populations. Lancet Diabetes Endocrinol 7(1):9–11. https://doi.org/10.1016/S2213-8587(18)30316-4
Dennis JM, Shields BM, Henley WE, Jones AG, Hattersley AT (2019) Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. Lancet Diabetes Endocrinol 7(6):442–451. https://doi.org/10.1016/S2213-8587(19)30087-7
Tanabe H, Saito H, Kudo A et al (2020) Factors associated with risk of diabetic complications in novel cluster-based diabetes subgroups: a Japanese retrospective cohort study. J Clin Med 9(7):2083. https://doi.org/10.3390/jcm9072083
Bennet L, Nilsson C, Mansour-Aly D, Christensson A, Groop L (2020) Ahlqvist E (2020) adult-onset diabetes in middle eastern immigrants to Sweden: novel subgroups and diabetic complications-the all new diabetes in Scania cohort diabetic complications and ethnicity. Diabetes Metab Res Rev 29:e3419. https://doi.org/10.1002/dmrr.3419
Fedotkina O, Sulaieva O, Ozgumus T et al (2021) Novel reclassification of adult diabetes is useful to distinguish stages of β-cell function linked to the risk of vascular complications: the DOLCE study from northern Ukraine. Front Genet 12:637945. https://doi.org/10.3389/fgene.2021.637945
Xing L, Peng F, Liang Q et al (2021) Clinical characteristics and risk of diabetic complications in data-driven clusters among type 2 diabetes. Front Endocrinol (Lausanne) 12:617628. https://doi.org/10.3389/fendo.2021.617628
Maalmi H, Herder C, Bönhof GJ et al (2021) Differences in the prevalence of erectile dysfunction between novel subgroups of recent-onset diabetes. Diabetologia. https://doi.org/10.1007/s00125-21-05607-z (in press)
Bonadonna RC, Trombetta M, Zusi C, Dauriz M, Boselli ML, Bonora E (2019) Novel subgroups of type 2 diabetes at diagnosis in a southern European cohort. Diabetes 68(Supplement 1):1502-P (abstract). https://doi.org/10.2337/db19-1502-P
Slieker RC, Donnelly LA, Fitipaldi H et al (2021) Replication and cross-validation of type 2 diabetes subtypes based on clinical variables: an IMI-RHAPSODY study. Diabetologia 64(9):1982–1989. https://doi.org/10.1007/s00125-021-05490-8
Bancks MP, Bertoni AG, Carnethon M et al (2021) Association of Diabetes Subgroups with Race/ethnicity, risk factor burden and complications: the MASALA and MESA studies. J Clin Endocrinol Metab 106(5):e2106–e2115. https://doi.org/10.1210/clinem/dgaa962
Bancks MP, Chen H, Balasubramanyam A et al (2021) Type 2 diabetes subgroups, risk for complications, and differential effects due to an intensive lifestyle intervention. Diabetes Care 44(5):1203–1210. https://doi.org/10.2337/dc20-2372
Bello-Chavolla OY, Bahena-López JP, Vargas-Vázquez A et al (2020) Clinical characterization of data-driven diabetes subgroups in Mexicans using a reproducible machine learning approach. BMJ Open Diabetes Res Care 8(1):e001550. https://doi.org/10.1136/bmjdrc-2020-001550
Carrillo-Larco RM, Castillo-Cara M, Anza-Ramirez C, Bernabé-Ortiz A (2021) Clusters of people with type 2 diabetes in the general population: unsupervised machine learning approach using national surveys in Latin America and the Caribbean. BMJ Open Diabetes Res Care 9(1):e001889. https://doi.org/10.1136/bmjdrc-2020-001889
Anjana RM, Baskar V, Nair ATN et al (2020) Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data-driven cluster analysis: the INSPIRED study. BMJ Open Diabetes Res Care 8(1):e001506. https://doi.org/10.1136/bmjdrc-2020-001506
Xiong XF, Yang Y, Wei L, Xiao Y, Li L, Sun L (2021) Identification of two novel subgroups in patients with diabetes mellitus and their association with clinical outcomes: a two-step cluster analysis. J Diabetes Investig 12(8):1346–1358. https://doi.org/10.1111/jdi.13494
Kahkoska AR, Geybels MS, Klein KR et al (2020) Validation of distinct type 2 diabetes clusters and their association with diabetes complications in the DEVOTE, LEADER and SUSTAIN-6 cardiovascular outcomes trials. Diabetes Obes Metab 22(9):1537–1547. https://doi.org/10.1111/dom.14063
Zaharia OP, Strassburger K, Knebel B et al (2020) Role of Patatin-like phospholipase domain-containing 3 gene for hepatic lipid content and insulin resistance in diabetes. Diabetes Care 43(9):2161–2168. https://doi.org/10.2337/dc20-0329
Zou H, Duan W, Zhang Z, Chen X, Lu P, Yu X (2020) The circulating ANGPTL8 levels show differences among novel subgroups of adult patients with diabetes and are associated with mortality in the subsequent 5 years. Sci Rep 10(1):12859. https://doi.org/10.1038/s41598-020-69091-y
Roden M, Shulman GI (2019) The integrative biology of type 2 diabetes. Nature 576(7785):51–60. https://doi.org/10.1038/s41586-019-1797-8
Herder C, Maalmi H, Strassburger K et al (2021) Differences in biomarkers of inflammation between novel subgroups of recent-onset diabetes. Diabetes 70(5):1198–1208. https://doi.org/10.2337/db20-1054
Ratter-Rieck JM, Maalmi H, Trenkamp S et al (2021) Leukocyte counts and T cell frequencies differ between novel subgroups of diabetes and associate with metabolic parameters and biomarkers of inflammation. Diabetes 30:db210364. https://doi.org/10.2337/db21-0364
Fasolino M, Goldman N, Wang W et al (2020) Genetic variation in type 1 diabetes reconfigures the 3D chromatin Organization of T Cells and Alters Gene Expression. Immunity 52(2):257–274.e11. https://doi.org/10.1016/j.immuni.2020.01.003
Gao P, Uzun Y, He B et al (2019) Risk variants disrupting enhancers of TH1 and TREG cells in type 1 diabetes. Proc Natl Acad Sci U S A 116(15):7581–7590. https://doi.org/10.1073/pnas.1815336116
Slieker RC, Donnelly LA, Fitipaldi H et al (2021) Distinct molecular signatures of clinical clusters in people with type 2 diabetes: an IMIRHAPSODY study. Diabetes 10:db201281. https://doi.org/10.2337/db20-1281
Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR (2009) Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 373(9682):2215–2221. https://doi.org/10.1016/S0140-6736(09)60619-X
Carstensen M, Herder C, Kivimäki M et al (2010) Accelerated increase in serum interleukin-1 receptor antagonist starts 6 years before diagnosis of type 2 diabetes: Whitehall II prospective cohort study. Diabetes 59(5):1222–1227. https://doi.org/10.2337/db09-1199
Tabák AG, Carstensen M, Witte DR et al (2012) Adiponectin trajectories before type 2 diabetes diagnosis: Whitehall II study. Diabetes Care 35(12):2540–2547. https://doi.org/10.2337/dc11-2263
Donath MY, Dinarello CA, Mandrup-Poulsen T (2019) Targeting innate immune mediators in type 1 and type 2 diabetes. Nat Rev Immunol 19(12):734–746. https://doi.org/10.1038/s41577-019-0213-9
Lawler PR, Bhatt DL, Godoy LC et al (2021) Targeting cardiovascular inflammation: next steps in clinical translation. Eur Heart J 42(1):113–131. https://doi.org/10.1093/eurheartj/ehaa099
Veelen A, Erazo-Tapia E, Oscarsson J, Schrauwen P (2021) Type 2 diabetes subgroups and potential medication strategies in relation to effects on insulin resistance and beta-cell function: a step toward personalised diabetes treatment? Mol Metab 46:101158. https://doi.org/10.1016/j.molmet.2020.101158
Perreault L, Skyler JS, Rosenstock J (2021) Novel therapies with precision mechanisms for type 2 diabetes mellitus. Nat Rev Endocrinol 17(6):364–377. https://doi.org/10.1038/s41574-021-00489-y
Dennis JM (2020) Precision medicine in type 2 diabetes: using individualized prediction models to optimize selection of treatment. Diabetes 69(10):2075–2085. https://doi.org/10.2337/dbi20-0002
Ahlqvist E, Prasad RB, Groop L (2020) Subtypes of type 2 diabetes determined from clinical parameters. Diabetes 69(10):2086–2093. https://doi.org/10.2337/dbi20-0001
Del Prato S (2019) Heterogeneity of diabetes: heralding the era of precision medicine. Lancet Diabetes Endocrinol 7(9):659–661. https://doi.org/10.1016/S2213-8587(19)30218-9
Schüssler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ et al (2019) A longitudinal big data approach for precision health. Nat Med 25(5):792–804. https://doi.org/10.1038/s41591-019-0414-6
Khoshnejat M, Kavousi K, Banaei-Moghaddam AM, Moosavi-Movahedi AA (2020) Unraveling the molecular heterogeneity in type 2 diabetes: a potential subtype discovery followed by metabolic modeling. BMC Med Genet 13(1):119. https://doi.org/10.1186/s12920-020-00767-0
Berry SE, Valdes AM, Drew DA et al (2020) Human postprandial responses to food and potential for precision nutrition. Nat Med 26(6):964–973. https://doi.org/10.1038/s41591-020-0934-0
Contrepois K, Wu S, Moneghetti KJ et al (2020) Molecular choreography of acute exercise. Cell 181(5):1112–1130.e16. https://doi.org/10.1016/j.cell.2020.04.043
Lee MMY, Ghouri N, McGuire DK, Rutter MK, Sattar N (2021) Meta-analyses of results from randomized outcome trials comparing cardiovascular effects of SGLT2is and GLP-1RAs in Asian versus white patients with and without type 2 diabetes. Diabetes Care 44(5):1236–1241. https://doi.org/10.2337/dc20-3007
Redondo MJ, Balasubramanyam A (2021) Towards an improved classification of type 2 diabetes: lessons from research into the heterogeneity of a complex disease. J Clin Endocrinol Metab 22:dgab545. https://doi.org/10.1210/clinem/dgab545
Fitipaldi H, McCarthy MI, Florez JC, Franks PW (2018) A global overview of precision medicine in type 2 diabetes. Diabetes 67(10):1911–1922. https://doi.org/10.2337/dbi17-0045
Lugner M, Gudbjörnsdottir S, Sattar N et al (2021) Comparison between data-driven clusters and models based on clinical features to predict outcomes in type 2 diabetes: nationwide observational study. Diabetologia 64(9):1973–1981. https://doi.org/10.1007/s00125-021-05485-5
Langenberg C, Sharp SJ, Franks PW et al (2014) Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med 11(5):e1001647. https://doi.org/10.1371/journal.pmed.1001647
Mahajan A, Taliun D, Thurner M et al (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50(11):1505–1513. https://doi.org/10.1038/s41588-018-0241-6
Authors’ relationships and activities
CH received a research grant from Sanofi-Aventis outside the submitted work. CH is a member of the editorial board of Diabetologia. MR received fees as a member of advisory boards or as a speaker from Allergan, Boehringer Ingelheim Pharma, Bristol-Myers Squibb, Eli Lilly, Fishawack Group, Gilead Sciences, Novartis Pharma, Intercept Pharma, Inventiva, Novo Nordisk, Target NASH and Terra Firma, and has been involved with clinical trial research for Boehringer Ingelheim, Danone Nutricia Research and Sanofi-Aventis, all outside the submitted work.
Funding
Open Access funding enabled and organized by Projekt DEAL. The German Diabetes Center (DDZ) is supported by the Ministry of Culture and Science of the State of North Rhine-Westphalia and the German Federal Ministry of Health. This work was supported in part by a grant from the German Federal Ministry of Education and Research to the German Center for Diabetes Research (DZD).
Author information
Authors and Affiliations
Contributions
Both authors were responsible for drafting the article and revising it critically for important intellectual content. Both authors approved the version to be published.
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Figure slide
(PPTX 230 kb)
ESM
(PDF 144 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Herder, C., Roden, M. A novel diabetes typology: towards precision diabetology from pathogenesis to treatment. Diabetologia 65, 1770–1781 (2022). https://doi.org/10.1007/s00125-021-05625-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-021-05625-x