Abstract
Aims/hypothesis
PARL, the gene encoding presenilins-associated rhomboid-like protein, maps to chromosome 3q27 within a quantitative trait locus that influences components of the metabolic syndrome. Recently, an amino acid substitution (Leu262Val, rs3732581) in PARL was associated with fasting plasma insulin levels in a US white population (N=1031). This variant was also found to modify the positive association between age and fasting insulin. The aim of this study was to test whether these findings could be replicated in two UK population-based cohorts.
Methods
Participants from the Medical Research Council Ely and Hertfordshire cohort studies were genotyped for this variant using a SNaPshot primer extension assay and Taqman assay respectively. Full phenotypic and genotypic data were available for 3,666 study participants.
Results
Based on a dominant model, we found no association between the Leu262Val polymorphism and fasting insulin levels (p=0.79) or BMI (p=0.98). We did not observe the previously reported interaction between age and genotype on fasting insulin (p=0.14).
Conclusions/interpretation
Despite having greater statistical power, our data do not support the previously reported association between PARL Leu262Val and fasting plasma insulin levels, a measure of insulin resistance. Our findings indicate that this variant is unlikely to be an important contributor to insulin resistance in UK populations.
Similar content being viewed by others
Introduction
The presenilins-associated rhomboid-like (PARL) protein (encoded by PARL) is a mitochondrial intramembrane protease conserved in eukaryotes and prokaryotes [1, 2]. In yeast, deletion of Rbd1/Pcp1 (the yeast homologue of PARL) leads to respiratory defects, impaired growth and small fragmented mitochondria. These phenotypes can be rescued by mammalian PARL, suggesting a conserved role in mitochondrial membrane remodelling [3, 4]. In Psammomys obesus, expression of the PARL homologue is reduced by 50% in skeletal muscle of obese, type 2 diabetic animals compared with lean glucose-tolerant animals. PARL expression is also negatively correlated with blood glucose and plasma insulin levels [5]. Consistent with these observations, in a Mexican American population, PARL expression was positively correlated with insulin sensitivity and citrate synthase activity, a marker of mitochondrial oxidative capacity [5]. Furthermore, PARL maps to chromosome 3q27 within a region that has been linked to quantitative traits that are components of the metabolic syndrome, such as BMI, waist and hip circumference, body weight, fasting plasma insulin and insulin:glucose ratio [6].
Based on its chromosomal position, and on animal and human data suggesting that PARL expression is associated with insulin sensitivity, variation at this gene was previously investigated for association with fasting plasma insulin and BMI in a US population. A common single nucleotide polymorphism (SNP) in exon 7 resulting in a leucine to valine substitution at amino acid residue 262 was genotyped in 1,031 US white individuals, a sub-group of the original population in which the quantitative trait locus was discovered [5, 6]. In a dominant model, PARL genotype was associated with fasting plasma insulin levels and modified the positive association between age and fasting insulin. There was no statistically significant association between this variant and BMI [5].
The aim of our study was to investigate whether the PARL Leu262Val genotype had a similar association with fasting plasma insulin levels in participants from two UK population-based cohorts.
Subjects and methods
Definition of cohorts
The Medical Research Council (MRC) Ely Study is a population-based cohort study of the aetiology and pathogenesis of type 2 diabetes and related metabolic disorders in the UK [7]. This analysis included 1,721 white Europid men and women aged 35 to 79 years without diagnosed diabetes. Participants underwent standard anthropometric measurements and a 75-g OGTT. Plasma glucose was measured using the hexokinase method.
Plasma-specific insulin was determined by two-site immunometric assays with either 125I or alkaline phosphatase labels. Cross-reactivity was <0.2% with intact proinsulin at 400 pmol/l and <1% with 32–33 split proinsulin at 400 pmol/l. Interassay CVs were 6.6% at 28.6 pmol/l (n=99), 4.8% at 153.1 pmol/l (n=102) and 6.0% at 436.7 pmol/l (n=99). Informed consent was obtained from all participants and ethical approval for the study was granted by the Cambridge Local Research Ethics Committee.
The Hertfordshire Cohort Study was recruited from the cohort of people born in Hertfordshire between 1931 and 1939. The cohort definition has been described previously [8]. Intact insulin was measured by in-house immunofluorimetric two-site assays (DELFIA system) based on published methods [9]. The CV for interassay imprecision ranged from 6 to 10% for low-, medium- and high-quality control samples for intact insulin and from 7 to 15% for insulin precursors. Glucose was assayed on an Advia 1650 autoanalyser (Bayer Diagnostics, Leverkusen, Germany). In this study we attempted to genotype 2,341 samples from North and East Hertfordshire.
Genotyping
DNA was available for 1,706 Ely cohort samples and 2,341 Hertfordshire cohort samples. The Leu262Val SNP (rs3732581) was genotyped in Ely using a multiplex kit according to the manufacturer’s instructions (ABI PRISM SNaPshot; Applied Biosystems, Foster City, CA, USA). Analysis was carried out using GeneMapper software (version 3.0; Applied Biosystems). The analysis included 120 water blanks, and 86 DNA samples (2.2%) were in duplicate on the plates. The genotyping success rate was 95%, with no discordance between replicate samples. Hertfordshire cohort samples were genotyped using a custom SNP assay (TaqMan; Applied Biosystems) on 10 ng of degenerate oligonucleotide primed DNA. Allele calling was performed on a sequence detection system (ABI PRISM 7900HT; Applied Biosystems). The analysis included 155 water blanks and 91 duplicates. The genotyping success rate was 96%. There was no discordance between replicate samples.
Statistical analysis
All analyses were carried out in Stata version 8 (StataCorp, College Station, TX, USA). Deviation of Leu262Val genotype from Hardy–Weinberg equilibrium was assessed using a χ 2 test. Only participants with genotype, fasting insulin and BMI data were included in the analysis, i.e. 1,608 Ely samples and 2,058 Hertfordshire samples (total=3,666). Linear regression analysis was used to assess the association between genotype and fasting plasma insulin or BMI in Ely and Hertfordshire cohorts separately. Based on the previous reported association, our primary analysis was for a dominant genetic model and we tested genotype–age interactions using log-likelihood ratio tests. In secondary exploratory analyses, we used regression analysis to assess the association between Leu262Val genotype and three further quantitative traits: fasting plasma glucose, 2-h glucose, and 30-min insulin increment (the difference between 30 min and fasting insulin concentrations divided by the 30-min glucose concentration) in an OGTT test. We also investigated whether the association between genotype and fasting insulin was modified by BMI. To increase power we undertook all analyses on both cohorts together including an indicator term for study.
Results
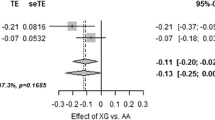
The PARL Leu262Val variant was in Hardy–Weinberg equilibrium in the combined Ely and Hertfordshire dataset (p=0.94). Demographic, anthropometric and biochemical characteristics according to Leu262Val genotype for the separate cohorts are shown in Table 1. We did not detect any statistically significant association between genotype and fasting plasma insulin, either in the Ely (p=0.49) or Hertfordshire (p=0.23) populations, or in the meta-analysis adjusting for cohort (p=0.79) (Table 2).
We found no association between genotype and BMI in Ely (p=0.65), Hertfordshire (p=0.75) or in the combined dataset (p=0.94). Furthermore, no age-dependent effects of genotype on fasting insulin levels were detected in either of the separate cohorts (p=0.40 for Ely, p=0.21 for Hertfordshire) or in our pooled analysis adjusted for study (p=0.14).
The statistically significant association between genotype and fasting insulin reported by the original study was based on a US cohort with higher mean BMI than our UK cohorts [5]. In an exploratory analysis, we investigated the impact of BMI on the association between genotype and fasting insulin in the pooled dataset. In an analysis restricted to individuals in the upper tertile for BMI (mean BMI=32.3 kg/m2, n=1,276), we found no statistically significant association between PARL genotype and fasting insulin (p=0.23). Second, we fitted a BMI–genotype interaction term, but this, too, was not significantly associated with fasting plasma insulin (p=0.88).
Finally, we assessed the association between this genetic variant and other quantitative metabolic traits (fasting glucose levels [mmol/l], 2-h glucose levels [mmol/l] and 30-min insulin increment [pmol/mmol]) with PARL genotype. No statistically significant associations were found (data not shown).
Discussion
Based on 3,666 study participants with full genotypic and phenotypic data, we found no evidence of an association between the PARL Leu262Val SNP and fasting insulin levels. Nor were we able to replicate the genotype–age interaction on fasting insulin levels [5], suggesting that this genetic variant is not an important contributor to variation in fasting insulin levels in UK populations.
Several reasons could explain the lack of agreement between our data and that previously published. The original study showed that plasma insulin levels were significantly higher in GG compared with GC/CC genotypic groups and that plasma insulin increased more rapidly with age in GG homozygotes [6]. The study population consisted of extended families recruited from the membership of the Take Off Pounds Sensibly organisation; mean BMI in this population was greater than in the two UK cohorts included in our analysis (33.4±7.2 kg/m2 in the US cohort compared with 27.3±4.5 kg/m2 in the UK cohorts). To examine the possibility that PARL Leu262Val is more strongly associated with fasting plasma insulin in more overweight people, we repeated our analyses in the restricted population of participants in the upper tertile of the distribution of BMI in our two cohorts. No statistically significant impact of genotype on fasting insulin was detected, nor was there evidence for an interaction between BMI and genotype on fasting insulin levels. These data indicate that failure to replicate the previous result is unlikely to be due to population differences in BMI.
The difference in results could also be explained by other genetic and/or environmental differences between the US and UK study populations. For example, the degree of linkage disequilibrium between the PARL SNP and the putative unmeasured true functional variants may vary between the cohorts. Alternatively, our study may have lacked the statistical power to detect the association between Leu262Val genotype and plasma insulin levels. However, we estimated that our pooled study would have had a 92% probability of detecting the previously reported effect size. The power of the previously reported study was lower due to the smaller sample size. In our stratified analyses, our power to detect an association of the previously reported effect size in the upper tertile of BMIs (n=1276) was 64%. We therefore cannot rule out a false negative finding in this stratified analysis.
We detected a nominally significant difference in age between those Ely subjects with a C allele and those without (Table 1), which is probably a chance finding resulting from multiple testing, as it is not apparent in the combined Ely and Hertfordshire dataset. Although there was a difference in average fasting plasma insulin between the Ely and Hertfordshire cohorts, most probably related to between-laboratory variation, this difference was not associated with genotype and would not have affected our results, as we included an adjustment for study cohort in our pooled analysis.
In summary, our results suggest that PARL Leu262Val is unlikely to be an important contributor to insulin resistance in UK populations. However, given the relationship between PARL expression levels and states of insulin resistance, this gene remains an interesting candidate in the development of type 2 diabetes and obesity. A wider range of SNPs could be typed in additional, larger, case-control and/or quantitative trait cohorts to elucidate the role of this gene in metabolic traits.
Abbreviations
- MRC:
-
Medical Research Council
- PARL:
-
presenilins-associated rhomboid-like
- SNP:
-
single nucleotide polymorphism
References
Pellegrini L, Passer BJ, Canelles M et al (2001) PAMP and PARL, two novel putative metalloproteases interacting with the COOH-terminus of Presenilin-1 and -2. J Alzheimers Dis 3:181–190
Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L (2003) The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol 4:R19
McQuibban GA, Saurya S, Freeman M (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423:537–541
Sik A, Passer BJ, Koonin EV, Pellegrini L (2004) Self-regulated cleavage of the mitochondrial intramembrane-cleaving protease PARL yields Pbeta, a nuclear-targeted peptide. J Biol Chem 279:15323–15329
Walder K, Kerr-Bayles L, Civitarese A et al (2005) The mitochondrial rhomboid protease PSARL is a new candidate gene for type 2 diabetes. Diabetologia 48:459–468
Kissebah AH, Sonnenberg GE, Myklebust J et al (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA 97:14478–14483
Wareham NJ, Hennings SJ, Byrne CD, Hales CN, Prentice AM, Day NE (1998) A quantitative analysis of the relationship between habitual energy expenditure, fitness and the metabolic cardiovascular syndrome. Br J Nutr 80:235–241
Syddall HE, Aihie Sayer A, Dennison EM, Martin HJ, Barker DJ, Cooper C (2005) Cohort profile: the Hertfordshire cohort study. Int J Epidemiol 34:1234–1242
Sobey WJ, Beer SF, Carrington CA et al (1989) Sensitive and specific two-site immunoradiometric assays for human insulin, proinsulin, 65–66 split and 32–33 split proinsulins. Biochem J 260:535–554
Acknowledgements
This work was supported in part by The Wellcome Trust (K. A. Fawcett, I. Barroso), the Medical Research Council (N. J. Wareham, S. O’Rahilly). I. Barroso and S. O’Rahilly also acknowledge funding from EU FP6 grant (contract no. LSHM-CT-2003-503041). The Hertfordshire population collections were undertaken by E. Dennison and A. Aihie Sayer. The Hertfordshire DNA banks were developed under MRC Programme Grant (G9828424) and Wessex Medical Trust (Hope)/National Lottery funding to I. N. M. Day. We acknowledge the technical assistance of M. Sims (MRC Epidemiology Unit, Strangeways Research Laboratory, Cambridge), T. Briggs (supported by the University of Southampton School of Medicine) and N. Graham. T. Gaunt, a BHF Intermediate Fellow, provided laboratory server database management.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fawcett, K.A., Wareham, N.J., Luan, J. et al. PARL Leu262Val is not associated with fasting insulin levels in UK populations. Diabetologia 49, 2649–2652 (2006). https://doi.org/10.1007/s00125-006-0443-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-006-0443-9