Abstract
Aims/hypothesis
This study aimed to identify genes that are expressed in skeletal muscle, encode proteins with functional significance in mitochondria, and are associated with type 2 diabetes.
Methods
We screened for differentially expressed genes in skeletal muscle of Psammomys obesus (Israeli sand rats), and prioritised these on the basis of genomic localisation and bioinformatics analysis for proteins with likely mitochondrial functions.
Results
We identified a mitochondrial intramembrane protease, known as presenilins-associated rhomboid-like protein (PSARL) that is associated with insulin resistance and type 2 diabetes. Expression of PSARL was reduced in skeletal muscle of diabetic Psammomys obesus, and restored after exercise training to successfully treat the diabetes. PSARL gene expression in human skeletal muscle was correlated with insulin sensitivity as assessed by glucose disposal during a hyperinsulinaemic–euglycaemic clamp. In 1,031 human subjects, an amino acid substitution (Leu262Val) in PSARL was associated with increased plasma insulin concentration, a key risk factor for diabetes. Furthermore, this variant interacted strongly with age to affect insulin levels, accounting for 5% of the variation in plasma insulin in elderly subjects.
Conclusions/interpretation
Variation in PSARL sequence and/or expression may be an important new risk factor for type 2 diabetes and other components of the metabolic syndrome.
Similar content being viewed by others
Introduction
Type 2 diabetes mellitus is a highly prevalent metabolic disease and a major cause of cardiovascular disease, kidney failure and blindness [1, 2]. Type 2 diabetes is characterised by insulin resistance and an insulin secretory defect. The role of skeletal muscle mitochondrial dysfunction in the development of insulin resistance and type 2 diabetes has received considerable attention recently. The evidence suggests that reduced expression and activity of enzymes involved in mitochondrial processes, including oxidative phosphorylation, the tricarboxylic acid cycle and beta-oxidation of fatty acids, are characteristic of insulin-resistant skeletal muscle [3–9]. These studies support findings that overall reductions in oxidative and phosphorylation capacity of mitochondria occur in insulin resistance and as part of the ageing process [10–15]. Indeed, there is a large body of evidence showing that mitochondrial functional capacity is a key component of the process of ageing, and of the development of a range of diseases. In insulin-resistant skeletal muscle, several studies have demonstrated reductions in the size and number of mitochondria, suggesting that defective mitochondrial biogenesis may be an important factor in the development of obesity and type 2 diabetes [7, 10, 12, 14, 16]. Interestingly, several studies have shown evidence of mitochondrial impairment in individuals with increased susceptibility (family history) to type 2 diabetes before the onset of symptoms [8, 9], suggesting that mitochondrial dysfunction may contribute to the development of type 2 diabetes.
There is also evidence to suggest that, by contributing to the accumulation of triglycerides in skeletal muscle, impaired mitochondrial beta-oxidation of fatty acids or dysregulation of the proportionality between glycolytic and oxidative activity could be a causal factor in the development of insulin resistance [4, 14, 17, 18]. This in turn could further impair the oxidative capacity of the muscle, generating a vicious cycle of increasing insulin resistance that eventually results in type 2 diabetes [4, 18].
In this study we screened skeletal muscle of Psammomys obesus (Israeli sand rats) for differentially expressed genes. P. obesus is a unique polygenic animal model of obesity and type 2 diabetes [19–21]. In their natural desert habitat, P. obesus subsist on a low-energy diet consisting mainly of Atriplex halimus (saltbush), and remain lean and free from diabetes. However, when housed under laboratory conditions and fed a standard rodent diet, some of the animals develop a range of metabolic disturbances including obesity, insulin resistance, dyslipidaemia and type 2 diabetes [19–21].
The aim of the current study was to identify genes that are expressed in skeletal muscle, encode proteins with functional significance in mitochondria, and are associated with type 2 diabetes.
Subjects, materials and methods
Experimental animals
A colony of P. obesus is maintained at Deakin University, Geelong, Australia. Breeding pairs are fed a diet of lucerne and chow, to which they have free access. Experimental animals were weaned at 4 weeks of age and given a diet of standard laboratory chow, in which 12% of energy was derived from fat, 63% from carbohydrate and 25% from protein (Barastoc, Pakenham, Vic, Australia). Animals were housed in a temperature-controlled room (22±1°C) with a 12/12-h light–dark cycle (light 06.00–18.00 hours). Animals were classified as having normal glucose tolerance (nGT), impaired glucose tolerance (IGT) or being type 2 diabetic at 16 weeks of age according to their blood glucose and plasma insulin concentrations as previously described [19–21]. For the exercise-training study, 15 male obese type 2 diabetic P. obesus underwent exercise training (running 1 km/day in 1 h) for 3 weeks. A separate group of eight age-matched, non-exercised male obese, type 2 diabetic P. obesus were included as controls.
Whole blood glucose was measured using an enzymatic glucose analyser (Model 27; Yellow Springs Instruments, Columbus, OH, USA). Plasma insulin concentrations were determined using a double-antibody solid phase radioimmunoassay (Phadeseph; Kabi Pharmacia Diagnostics, Sweden). All experimental animals and procedures were maintained in accordance with the Code of Practice outlined by the National Health and Medical Research Council (NHMRC) and approved by the Deakin University Ethics Committee.
Membrane microarray
At 18 weeks of age, animals (n=5 in each group; lean nGT and obese type 2 diabetic) were killed and gastrocnemius muscles immediately removed, frozen in liquid nitrogen and stored at −80°C. RNA was isolated using TriZol (Invitrogen, Carlsbad, CA, USA). A Research Genetics Human GF 201 gene filter containing 5,568 clones (Research Genetics, Huntsville, AL, USA) was used to screen genes differentially expressed in the skeletal muscle of P. obesus according to the manufacturer’s instructions. Briefly, RNA from both groups was separately pooled (2 μg) and reverse transcribed in the presence of 7.4 MBq of [33P]dCTP. The membrane was hybridised, washed, and exposed to a phosphor-imager screen (Molecular Dynamics, Sunny Vale, CA, USA). The experiment was repeated twice, and averaged images were analysed and gene expression quantitated using Pathways V 2 software (Research Genetics). There are clearly limitations when using human arrays to screen for expression of P. obesus genes. However it should be noted that this was simply a preliminary screen to identify candidates for further analysis using Taqman PCR. At the time of this screen, no P. obesus arrays were available.
Bioinformatics
Sequences of genes differentially expressed according to the microarray experiments were analysed using publicly available software tools and databases including BLAST, ClustalW and MitoProt http://www.expasy.org, http://www.ncbi.nlm.nih.gov).
Gene expression of AGT-203 (Presenilens-associated rhomboid-like protein [PSARL] homologue) in P. obesus
The level of gene expression was quantified using real-time quantitative RT-PCR on an ABI Prism 7700-sequence detector. Cyclophilin (a house-keeping gene) was used to normalise the amount of cDNA in reactions. The primer sequences were as follows: AGT-203 forward, 5′-ccc acc tct gga aga aac tgt ct-3′; AGT-203 reverse 5′-cgg aca ctt ggg ttg tca ctt ct-3′; cyclophilin forward, 5′-cct tgc tgg tga tgg tgg aga a-3′; cyclophilin reverse 5′-gac ctg gcg ttt taa ctc aag-3′. Fluorogenic probe sequences were 5′-tta tcc ttc ccc cta ccc tat aag aac ttt gtg-3′ for AGT-203 and 5′-atc aac ctc ttc tgg cga cgg tca gg-3′ for cyclophilin. The AGT-203 and cyclophilin probes had the reporter dye FAM attached to the 5′ end and the quencher dye TAMRA attached to the 3′ end. PCR conditions consisted of a two-step PCR. Step 1 included a 2-min stage at 50°C and another at 95°C for 10 min to activate AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA, USA). Step 2 consisted of a denaturing step at 95°C for 15 s and an extension stage at 60°C for 1 min for 40 cycles.
Human subjects for PSARL gene expression analysis
Two groups of Mexican American volunteers matched for age and body composition were recruited at the General Clinical Research Center of the South Texas Veterans Health Care System in San Antonio. Eight of them were healthy normoglycaemic subjects with at least two first-degree relatives with type 2 diabetes (family history +; three men/five women; 36±3 years; 28±1 kg/m2), whereas the ten others were control subjects with no family history of type 2 diabetes (family history −; six men/four women; 37±4 years; 28±2 kg/m2). Insulin sensitivity was measured by a hyperinsulinaemic–euglycaemic clamp (40 mU·m−2·min−1), with infusion of [3H]-glucose lower in the family history positive subjects than the family history negative subjects (3.0±0.3 vs. 5.6±0.5 mg·kg−1·min−1; p=0.001). A percutaneous vastus lateralis muscle biopsy was performed and PSARL gene expression was measured using real-time quantitative RT-PCR. Primer sequences were as follows: PSARL forward, 5′-ttc tct gca gcg gac aat gat-3′; PSARL reverse 5′-tga cag caa cat tgg aga aca aa-3′. The fluorogenic probe sequence was 5′-ttt cac atc gaa tcc agc ctc aaa ggt c-3′. The Institutional Review Board of the University of Texas Health Science Center at San Antonio approved this research and all subjects gave written informed consent.
Human subject selection for association study
Human samples and phenotypic data were utilised from the Metabolic Risk Complications of Obesity Genes project. Details of the sample are discussed elsewhere, where the original localisation of a quantitative trait locus (QTL) influencing metabolic syndrome phenotypes to chromosomal region 3q27 is also described [22]. DNA samples for these individuals were used to genotype the publicly available PSARL exon 7 leucine→valine polymorphism (NCBI SNP Cluster ID rs3732581, details available at http://www.ncbi.nlm.nih.gov). Henceforth this is referred to as the Leu262Val polymorphism of PSARL, as it encodes a leucine to valine amino acid substitution at position 262 of the PSARL protein. These families were recruited from the Take Off Pounds Sensibly (TOPS) membership. TOPS provided mailing material on membership attending its chapters in ten states (Wisconsin, Illinois, Michigan, Iowa, Minnesota, Ohio, West Virginia, Missouri, Kentucky and Indiana). Families with at least two obese siblings (BMI >30 kg/m2), availability of one (or preferably both) parent(s), as well as at least one never-obese sibling and/or parent (BMI <27 kg/m2) were identified and contacted for ascertainment. Families were scheduled to visit satellite centres (four to six per state), where an experienced team undertook the phenotypic procedures. Informed consent was obtained from all subjects in this study, and all research protocols were approved by the Institutional Review Board of the Medical College of Wisconsin.
The data used in the present examination are taken from 1,031 individuals in 169 families of predominantly northern European ancestry and residing in the United States. This represents a subset of the original study. From the original 507 families, we chose the largest families since pedigree size and complexity correlates strongly with power to localise and identify QTLs [23]. Table 1 shows the pair-wise relationships represented by this data set.
Phenotypes
Detailed phenotypic information was available on these individuals, although for the present study we considered only two phenotypes, fasting plasma insulin concentrations and BMI. These phenotypes were chosen because they both exhibited log odds ratio (LOD) scores of greater than 3 for the region of the PSARL gene (chromosome 3q27; see Fig. 1). Additionally, they each represent one of the two main linkage peaks that were seen in the original genome-wide linkage scan in this population [22]. Standard anthropometric measures were obtained according to the World Health Organization recommendations with participants wearing light clothing. A double-antibody equilibrium radioimmunoassay (Linco Research, St. Charles, MO, USA) was used for the measurement of plasma insulin, using an antibody specific to human insulin. Quality controls were performed to ensure stability and reliability of the assays. Five pool sera of increasing peptide concentrations were used to evaluate the intra-assay and interassay coefficients of variance.
Location of the gene encoding PSARL in relation to evidence for linkage to obesity and diabetes traits in the TOPS study [22]
Genotyping
All genotyping was performed by mass spectrometry using the MassArray System (Sequenom, San Diego, CA, USA) according to the manufacturer’s instructions. Full details of the genotyping methodology are available from the authors.
Statistical genetic methods
In order to assess the influence of the PSARL Leu262Val polymorphism on insulin levels and BMI, we utilised a measured genotype approach allowing for the non-independence of phenotypes among family members [24, 25]. Measured genotype analysis is a form of association analysis that employs the genotypic information in a fixed effect model for the mean effects. Thus, it can be used to test the hypothesis that phenotypic means differ amongst genotypes. Because these traits were non-normally distributed, we employed a robust method for estimating parameters using a multivariate t-distribution [26]. Focal phenotypes (insulin and BMI) were adjusted for a number of covariates (smoking, menopausal status, exogenous sex hormones, sex, age) prior to genetic analysis as described elsewhere [22].
All parameter estimation was done by maximum likelihood under the assumption of a multivariate t-distribution using the computer package, SOLAR [27]. A formal test of association was obtained by calculating a robust likelihood ratio test statistic comparing a model in which the genotypic means were held equal against a model in which the genotypic means could vary. Because of prior evidence for an important mitochondrial function for PSARL and the cumulative damage to mitochondria seen during the ageing process, we also allowed for genotype-by-age interaction as previously described [28]. Briefly, genotype-by-age interaction requires the differential response of genotypes to the ageing process. Thus, the presence of different age trajectories as a function of genotype is de facto evidence for genotype-by-age interaction. Formal likelihood ratio tests were constructed to test whether the regressions of the focal phenotypes on age varied significantly across genotypic classes.
Basic statistical analyses
Comparisons between group mean values were performed using an unpaired Student’s t-test for two-group comparisons. For comparisons involving three or more groups, one-way ANOVA was performed using the LSD post hoc test for multiple comparisons.
Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS, Version 10.1; SPSS, Fullerton, CA, USA). Statistical significance was defined as p<0.05. All values are presented as means±SEM.
Results
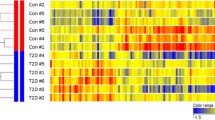
We used membrane microarray analysis to screen for genes with evidence of differential expression in skeletal muscle of lean, nGT and obese, type 2 diabetic P. obesus (n=5 in each group; body weight 178±3 vs 245±6 g, p<0.01; blood glucose 3.5±0.3 vs 16.7±1.9 mmol/l, p<0.01; plasma insulin 66±13 vs 746±199 μU/ml, p<0.01). A total of 54 genes showed preliminary evidence of differential expression (>2.0-fold ratio) between the muscle samples, of which 19 were known genes, and 35 were expressed sequence tags and were considered candidates for further analysis using Taqman PCR. The chromosomal locations of all 54 genes were determined by database searching and mapped on our in-house genomics database. Several of these differentially expressed genes encoded known mitochondrial proteins (for example SLC25A25, ATP5D and NDUFS5 all had reduced gene expression in muscle of diabetic P. obesus). However, none of these genes was located in genomic regions thought to contain diabetes susceptibility genes. Therefore we focused on one gene, which we named AGT-203 and which was expressed at 70% lower levels in the muscle of diabetic animals and was mapped to chromosome 3q27 in a region with strong evidence for linkage with obesity and type 2 diabetes [22, 29–34].
Sequence analysis revealed that AGT-203 was the P. obesus homologue (91% similarity) of PSARL [35]. The entire P. obesus PSARL sequence was determined by RACE, and the highly conserved amino acid alignment with other species is shown in Fig. 2. P. obesus PSARL mRNA is 1,295 bp in length, with an open reading frame of 1,128 nucleotides encoding a 376 amino acid protein. PSARL is a 42-M r protein with seven predicted transmembrane helices and a conserved rhomboid domain. Using the Mitoprot algorithm [36], PSARL was predicted to be exported to the mitochondria (probability 0.6033) with a putative cleavage sequence of MAWRGWAQRG (amino acids 1–10).
PSARL gene expression was measured in skeletal muscle of P. obesus using real-time quantitative RT-PCR. As shown in Fig. 3, the expression of the PSARL gene was reduced by 50% in skeletal muscle of obese, type 2 diabetic P. obesus relative to the lean, nGT animals (p<0.05). PSARL gene expression in the IGT animals lay between the nGT and type 2 diabetic groups (Fig. 3). Furthermore, the expression of PSARL was negatively correlated with blood glucose (r=−0.48, p<0.05) and plasma insulin (r=−0.51, p<0.01) concentrations. The expression of PSARL in gastrocnemius muscle was increased by 49% (p=0.036) in obese, type 2 diabetic P. obesus that were exercise-trained for 3 weeks, a procedure that reduced blood glucose (by 23%, p=0.038) and plasma insulin (by 36%, p=0.002), indicating improved insulin sensitivity (Fig. 4).
Effects of exercise training on (a) blood glucose, (b) plasma insulin and (c) gene expression of PSARL in gastrocnemius muscle of exercise-trained and control obese, type 2 diabetic P. obesus (n=12 in each group). Open columns, day 0; filled columns, day 21. PSARL gene expression was measured by Taqman PCR and is expressed relative to cyclophilin. *p<0.05 compared with day 0; # p<0.05 compared with control group
We also examined PSARL gene expression in skeletal muscle of 18 Mexican American subjects with and without a family history of type 2 diabetes. The skeletal muscle expression of the PSARL gene was similar in subjects with or without family history of type 2 diabetes (1.1±0.1 vs 1.1±0.2 arbitrary units in family history positive vs family history negative subjects, respectively). However, when only subjects with no family history of diabetes were considered, there was a positive correlation between the gene expression of PSARL and insulin sensitivity as assessed by glucose disposal during the hyperinsulinaemic–euglycaemic clamp (men; r=0.79, p=0.006; Fig. 5). Furthermore, in these subjects PSARL gene expression in skeletal muscle was correlated with the expression of citrate synthase, a marker of mitochondrial oxidative capacity (r=0.86, p<0.001). Although the numbers are small in this study and the data need to be interpreted with caution due to the possibility of type 1 error, we believe that these results support a potential association between PSARL and mitochondrial function and insulin sensitivity.
PSARL is located on chromosome 3q27 in a region with substantial evidence of linkage with metabolic syndrome phenotypes. PSARL lies only 2.5 Mb to the p-ter side of the linkage peak (approximately 202 cM on the genetic map used). The 1-LOD support interval for this QTL spans 10.8 Mb (192–227 on the genetic map). We therefore genotyped the common Leu262Val variant of PSARL in a cohort of 1,031 human subjects in which the QTL was originally discovered. The genomic structure of PSARL and the location of the Leu262Val variant are depicted in Fig. 6. The average age of the sample was 47.2 years, and included 759 women and 272 men. The average plasma insulin value (age-corrected) was 86.9 pmol/l, with similar means in women and men. The observed phenotypic standard deviation was 37.5 pmol/l. The observed heritability for insulin levels was 0.207 (p=0.0001). The average BMI (age corrected) for men was 30.2 kg/m2. The BMI of the women was on average 4.4 units higher. The observed phenotypic SD was 7.2 kg/m2. The heritability of BMI in this sample was 0.291 (p=0.0000001).
The Leu262Val polymorphism is due to a substitution of a C for a G, leading to a change in amino acid from a leucine to a valine. The frequency of the rarer C variant was 0.483. The genotypic distribution was consistent with Hardy–Weinberg equilibrium after taking into account family structure. Of the 1,031 individuals with complete data, 274 (26.6%) were GG homozygotes, 523 (50.7%) were GC heterozygotes and 234 (22.7%) were CC homozygotes.
Robust measured genotype analysis revealed a significant influence of this polymorphism on plasma insulin concentrations. The best fitting and most parsimonious model of gene action suggested dominance of the C allele so that genotypic classes for comparison included GG versus GC+CC genotypes. The more general model allowing the three possible genotypes to exhibit their own mean genotypic effects converged to the dominant model. When age was held constant at the average age of the population, GG homozygotes exhibited a mean plasma insulin that was 7.84±3.03 pmol/l higher (p=0.0098) than that observed for the GC+CC genotypic class. In addition, there was striking evidence of genotype-by-age interaction. GG homozygotes showed significantly greater slope with regard to age than did the GC+CC genotypic class. Plasma insulin concentrations increased substantially more rapidly with age in GG individuals (a slope difference of 0.313±0.097 pmol·l−1·year−1). A robust likelihood ratio test provided a p value of 8.6×10−5 for the test of genotype-by-age interaction.
A combined robust likelihood ratio test of the influence of the polymorphism on plasma insulin yielded a highly significant p value of 0.00025. This strongly supports the hypothesis that the Leu262Val polymorphism is either directly involved or correlated with another causal variant to influence the distribution of plasma insulin concentrations in this population.
Figure 7 depicts the pattern of genotype-by-age interaction graphically. The graph shows the model-predicted mean plasma insulin as a function of genotype. Clearly, plasma insulin concentrations in GG individuals increase substantially more rapidly with age than in individuals with the GC and CC genotypes.
The relative importance of this polymorphism on plasma insulin concentrations is thus a function of age. The increased difference between the genotypic means at higher ages indicates that additional genetic variance is expressed. This has an effect on the total heritability of the trait and the heritability that is specifically due to the polymorphism. Figure 8 shows the pattern of relative genetic variance in plasma insulin levels with age. At early ages, the polymorphism accounts for relatively little of the phenotypic variation. However, at older ages the polymorphism accounts for 5% of the total variation. This represents a substantial causal component of plasma insulin variation.
In contrast to the results for plasma insulin, the Leu262Val polymorphism did not appear to influence BMI. Using a model of gene action similar to that for plasma insulin concentrations, the robust likelihood ratio test was not significant (p=0.579).
Discussion
In these studies we have shown that PSARL, a mitochondrial intramembrane protease, is a new candidate gene for obesity and type 2 diabetes. We screened for genes with evidence of differential expression in skeletal muscle of lean, nGT and obese, type 2 diabetic P. obesus, and identified a total of 54 candidate genes for further investigation. The characteristics of these genes and their protein products were examined in detail using information available from public databases to identify those likely to have a functional role in mitochondria. This analysis highlighted one gene, AGT-203, which was the P. obesus homologue of human PSARL [35] and yeast Pcp1 [37, 38]. While the subcellular localisation of PSARL was not known at that time, Pcp1 contained a signature motif for mitochondrial localisation, and had been shown to interact with proteins known to reside within the mitochondria. Furthermore, bioinformatics analysis predicted a mitochondrial localisation for PSARL. Gene expression studies in P. obesus supported an association with insulin resistance, and in human subjects, PSARL gene expression in skeletal muscle correlated with both citrate synthase expression, a marker of mitochondrial oxidative capacity, and insulin sensitivity assessed by hyperinsulinaemic–euglycaemic clamp.
The rhomboid family of proteins, of which PSARL is a member, shows remarkable evolutionary conservation. They are a family of intramembrane serine proteases that have been found in almost all species of archaea, bacteria and eukaryotes [39, 40]. Rhomboids participate in regulated intramembrane proteolysis, a recently discovered process, whereby signalling proteins are cleaved at intracellular membranes to release active fragments in signal transduction cascades [41, 42].
Two recent studies demonstrated that the yeast homologue of PSARL (known as Rbd1, Pcp1 or Ygr101w) plays a key role in mitochondrial membrane remodelling [37, 38]. Mitochondrial structure and function are highly dependent on the processes of fusion and fission. Both of these studies demonstrated that processing of Mgm1, a dynamin-like GTP-ase, by Rbd1/Pcp1 was critical for the maintenance of mitochondrial structure and function [37, 38]. Yeast strains lacking functional Rbd1/Pcp1 exhibited respiratory defects and impaired growth, and had small, fragmented mitochondria [37]. Replacement with PSARL in Rbd1/Pcp1-deficient yeast restored Mgm1 processing and rescued growth rate and mitochondrial morphology [37]. These results demonstrated the importance of the yeast homologue of PSARL in mitochondrial function, and suggested that PSARL may play a similar role in mammalian physiology. Furthermore, PSARL was localised to mitochondria in COS cells [37].
The role of mitochondrial function in a range of diseases including obesity and type 2 diabetes is well known. Numerous studies have demonstrated defects in oxidative pathways in skeletal muscle of subjects with obesity and/or type 2 diabetes [3, 4, 7, 10, 11, 13], and it has been proposed that impaired bioenergetic capacity of skeletal muscle mitochondria is a key factor in the development of these diseases [7]. Furthermore, reduced mitochondrial function is a hallmark of the ageing process [14, 15], and could be a determining factor in the development of insulin resistance and other disorders related with ageing, e.g. neurodegenerative diseases.
Given the likely role of PSARL in maintaining mitochondrial membrane integrity and function, and the known defects of mitochondria in diabetes [3, 4, 7, 10, 11, 13], we suggest that altered activity of PSARL in skeletal muscle may be an important contributing factor in the development of insulin resistance and type 2 diabetes. Furthermore, the PSARL gene is located on chromosome 3q27, in a region that has been linked to obesity and type 2 diabetes in numerous genome-wide linkage scans.
To test for possible association between variation in the PSARL gene and obesity/type 2 diabetes, we genotyped the common C→G SNP in exon 7 of PSARL that encodes an amino acid substitution from leucine to valine (Leu262Val). The Leu262Val variant was significantly associated with plasma insulin concentration (p=0.0098), a proxy marker of insulin resistance. Additionally, there was strong evidence that this variant interacted with age to influence plasma insulin (p=8.6×10−5). The interaction between genotype and age was such that plasma insulin increased at a greater rate in individuals with the GG genotype than in those with the CG/CC genotypes. In older subjects, the Leu262Val profoundly affects plasma insulin concentration accounting for 5% of the variation, which is substantially more than the effects of other known covariates such as sex, menopausal status, smoking, and even diabetes status in this population. The strong evidence for differential expression of genetic variance as a function of age points to the potential importance of age-related differences as a general phenomenon in the phenotypic dimensions of common diseases such as type 2 diabetes.
The chromosome 3q QTL that influences metabolic syndrome phenotypes (including obesity and type 2 diabetes) is one of the most replicated human quantitative trait linkages [22, 29–34]. Given the relatively low power of human genetic studies for mapping QTLs, this remarkable pattern of replicated QTL localisation is strongly suggestive of either multiple gene involvement in the region of 3q27 or variants within a single gene with very large effects. The former hypothesis appears to be the most likely. PSARL appears to be one of the loci contributing to the chromosome 3 QTL cluster. Interestingly, it affects plasma insulin concentration but not BMI suggesting that there are multiple components in the QTL cluster. Much recent attention has been given to an obvious candidate gene in this region, APM1, which encodes for adiponectin, and multiple association studies have found weak associations with variations in this gene and diabetes/obesity-related traits [43]. APM1 appears to represent one of the contributing genes to this metabolic syndrome QTL. The data we have presented in this communication add the PSARL gene as another genetic component contributing to this QTL.
The interaction between genotype at Leu262Val of PSARL and age, and the association with plasma insulin concentration, is consistent with the hypothesis that defects in mitochondria tend to accumulate throughout life resulting in gradual diminution of overall mitochondrial function [14, 15]. The Leu262Val variant is a conservative amino acid substitution in the fourth transmembrane helix of PSARL. While this substitution does not impact directly on the predicted catalytic sites of PSARL, it could be reasonably expected to alter the conformation of the protein in the mitochondrial membrane, and may affect its activity. This mitochondrial impairment is likely to contribute to the development of insulin resistance over time, as we demonstrated in our genotype-by-age interaction with plasma insulin concentration for this variant.
In summary, we have shown that PSARL expression in skeletal muscle is reduced in type 2 diabetes in P. obesus, and that this can be alleviated, along with insulin resistance, by exercise training. In human subjects, PSARL gene expression in skeletal muscle was associated with mitochondrial oxidative capacity and insulin sensitivity, providing further support for the contention that PSARL plays a role in the development of insulin resistance through its effects on mitochondrial function. Furthermore, genetic variation at Leu262Val of PSARL was associated with insulin resistance in an age-dependent manner. Impaired mitochondrial function is a key factor in the development of insulin resistance, as well as many other diseases, particularly those associated with ageing. As insulin resistance develops, PSARL expression/activity and mitochondrial function probably decline further, leading to a vicious cycle that eventually results in type 2 diabetes. We suggest that variation in the expression and activity of PSARL is an important new risk factor for the development of insulin resistance and type 2 diabetes.
Abbreviations
- LOD:
-
Log odds ratio
- nGT:
-
Normal glucose tolerant
- PSARL:
-
Presenilins-associated rhomboid-like protein
- QTL:
-
Quantitative trait locus
- TOPS:
-
Take off pounds sensibly
References
Zimmet P (2000) Globalization, coca-colonization and the chronic disease epidemic: can the doomsday scenario be averted? J Intern Med 247:301–310
Zimmet P, Alberti KGMM, Shaw J (2001) Global and societal implications of the diabetes epidemic. Nature 414:782–787
Vondra K, Rath R, Bass A, Slabochova Z, Teisinger J, Vitek V (1977) Enzyme activities in quadriceps femoris muscle of obese diabetic male patients. Diabetologia 13:527–529
Simoneau J-A, Kelley DE (1997) Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol 83:166–171
Yang X, Pratley RE, Tokraks S, Bogardus C, Permana PA (2002) Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia 45:1584–1593
Sreekumar R, Halvatsiosis P, Schimke JC, Nair KS (2002) Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 51:1913–1920
Kelley DE, He J, Menshikova EV, Ritov VB (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51:2944–2950
Patti ME, Butte AJ, Crunkhorn S et al (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF-1. Proc Natl Acad Sci U S A 100:8466–8471
Mootha VK, Lindgren CM, Eriksson KF et al (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273
Bjorntorp P, Schersten T, Fagerberg SE (1967) Respiration and phosphorylation of mitochondria isolated from the skeletal muscle of diabetic and normal subjects. Diabetologia 3:346–352
Simoneau J-A, Colberg SR, Thaete FL, Kelley DE (1995) Skeletal muscle glycolytic and oxidative enzyme capacities are determinants of insulin sensitivity and muscle composition in obese women. FASEB J 9:273–278
Antonetti DA, Reynet C, Kahn CR (1995) Increased expression of mitochondrial-encoded genes in skeletal muscle of humans with diabetes mellitus. J Clin Invest 95:1383–1388
Simoneau J-A, Veerkamp JH, Turcotte LP, Kelley DE (1999) Markers of capacity to utilize fatty acids in human skeletal muscle: relation to insulin resistance and obesity and effects of weight loss. FASEB J 13:2051–2060
Petersen KF, Befroy D, Dufour S et al (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300:1140–1142
Rooyackers OE, Adey DB, Ades PA, Nair KS (1996) Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc Natl Acad Sci U S A 93:15364–15369
Song J, Oh JY, Sung YA, Pak YK, Park KS, Lee HK (2001) Peripheral blood mitochondrial DNA content is related to insulin sensitivity in offspring of type 2 diabetic patients. Diabetes Care 24:865–869
Kelley DE, Simoneau J-A (1994) Impaired free fatty acid utilization by skeletal muscle in non-insulin-dependent diabetes mellitus. J Clin Invest 94:2349–2356
Kelley DE, Mandarino LJ (2000) Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes 49:677–683
Barnett M, Collier GR, Collier FM, Zimmet P, O’Dea K (1994) A cross-sectional and short-term longitudinal characterisation of NIDDM in P. obesus. Diabetologia 37:671–676
Collier GR, McMillan JS, Windmill K et al (2000) Beacon: a novel gene involved in the regulation of energy balance. Diabetes 49:1766–1771
Walder K, Kantham L, McMillan JS et al (2002) Tanis: a link between type 2 diabetes and inflammation? Diabetes 51:1859–1866
Kissebah AH, Sonnenberg GE, Myklebust J et al (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci U S A 97:14478–14483
Blangero J, Williams JT, Almasy L (2003) Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost 1:1391–1397
Hopper JL, Mathews JD (1982) Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet 46:373–383
Boerwinkle E, Chakraborty R, Sing CF (1986) The use of measured genotype information in the analysis of quantitative phenotypes in man: I. Models and analytical methods. Ann Hum Genet 50:181–194
Lange K, Little RJA, Taylor JMG (1989) Robust statistical modelling using the multi-variate T-distribution. J Acoust Soc Am 84:881–896
Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211
Blangero J (1993) Statistical genetic approaches to human adaptability. Hum Biol 65:941–966
Francke S, Manraj M, Lacquemant C et al (2001) A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 10:2751–2765
Luke A, Wu X, Zhu X, Kan D, Su Y, Cooper R (2003) Linkage for BMI at 3q27 region confirmed in an African-American population. Diabetes 52:1284–1287
Mori Y, Otabe S, Dina C et al (2002) Genome-wide search for type 2 diabetes in Japanese affected sib-pairs confirms susceptibility genes on 3q, 15q, and 20q and identifies two new candidate loci on 7p and 11p. Diabetes 51:1247–1255
Wu X, Cooper RS, Borecki I et al (2002) A combined analysis of genomewide linkage scans for body mass index from the National Heart, Lung, and Blood Institute Family Blood Pressure Program. Am J Hum Genet 70:1247–1256
Vionnet N, Hani E-H, Dupont S et al (2000) Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21–q24. Am J Hum Genet 67:1470–1480
Zhu X, Cooper RS, Luke A et al (2002) A genome-wide scan for obesity in African-Americans. Diabetes 51:541–544
Pellegrini L, Passer BJ, Canelles M et al (2001) PAMP and PSARL, two novel putative metalloproteases interacting with the COOH-terminus of Presenilin-1 and -2. J Alzheimer’s Dis 3:181–190
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
McQuibban GA, Saurya S, Freeman M (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423:537–541
Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem 278:27781–27788
Urban S, Lee JR, Freeman M (2001) Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107:173–182
Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier M-C, Pellegrini L (2003) The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol 4:R19
Brown MS, Ye J, Rawson RB, Goldstein JL (2003) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391–398
Golde TE, Eckman CB (2003) Physiologic and pathologic events mediated by intramembranous and juxtamembranous proteolysis. Sci Signal Transduct Knowl Environ 2003:RE4
Vasseur F, Lepretre F, Lacquemant C, Froguel P (2003) The genetics of adiponectin. Curr Diabetes Rep 3:151–158
Acknowledgements
Funding for this study was provided by ChemGenex Pharmaceuticals, Australia. The human genetics work was supported by Grants HL34989, DK54026, MH59490, and RR00058 from the National Institutes of Health. The genotyping was undertaken under the auspices of the Mammalian Genotyping Service of the National Institutes of Health, funds being allocated to the Marshfield Medical Research Foundation. TOPS provided funds for establishment of the family database, phenotyping, and linkage analysis. Part of the phenotyping costs was also provided through a collaborative research agreement with Millennium Pharmaceuticals.
Duality of interest: K. Walder, L. Kerr-Bayles and N. Bishara are employed by Deakin University which is currently in receipt of a grant from ChemGenex Pharmaceuticals (formerly AGT Bioscience) to investigate the genetic basis of type 2 diabetes, obesity and related metabolic disorders. P. Zimmet, J. Jowett, J. Curran and K. Elliott are employed by the International Diabetes Institute, which is currently in receipt of a research grant from ChemGenex Pharmaceuticals to investigate the genetic basis of type 2 diabetes, obesity and related metabolic disorders. G. Collier is the Chief Executive Officer of ChemGenex Pharmaceuticals. P. Zimmet chairs the Scientific Advisory Board of ChemGenex Pharmaceuticals and hold shares in the company. K. Walder holds a part-time position as a consultant for ChemGenex Pharmaceuticals, and J. Blangero is a member of the Scientific Advisory Board for ChemGenex Pharmaceuticals. J. Jowett, J. Curran and K. Elliott are holders of unexercised options for shares in ChemGenex Pharmaceuticals.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Walder, K., Kerr-Bayles, L., Civitarese, A. et al. The mitochondrial rhomboid protease PSARL is a new candidate gene for type 2 diabetes. Diabetologia 48, 459–468 (2005). https://doi.org/10.1007/s00125-005-1675-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-005-1675-9