Abstract
Aims/hypothesis
Plasma cell membrane glycoprotein 1 (PC-1) inhibits insulin signalling by direct interaction with the insulin receptor α subunit. This inhibition is enhanced by the minor Q allele of the K121Q polymorphism (rs1044498) in the gene (ENPP1) encoding PC-1. This polymorphism has been studied in relation to insulin resistance, type 2 diabetes and obesity in several populations with conflicting results. We assessed the impact of the ENPP1 K121Q polymorphism on type 2 diabetes, obesity and quantitative metabolic traits in 7,333 Danes.
Subjects and methods
The K121Q polymorphism was genotyped in the population-based Inter99 study cohort (5,961 subjects) and in a group of 1,386 patients with type 2 diabetes. All subjects were Danish whites.
Results
No significant associations with type 2 diabetes or related quantitative metabolic traits, including measures of insulin resistance, were detected. However, a meta-analysis of the present and published studies revealed an association with type 2 diabetes (odds ratio per Q allele, 1.17 [95% CI 1.10–1.25], p=1×10−6). In case–control studies comparing subjects of different BMI strata, we observed a putative association of the codon 121 QQ genotype with being overweight (BMI>25 kg/m2; odds ratio 1.63 [95% CI 1.09–2.46], p=0.015), an association not observed when comparing other levels of BMI or when analysing BMI as a quantitative trait.
Conclusions/interpretation
In a meta-analysis, the ENPP1 codon 121 Q allele associates with type 2 diabetes. However, a similar association was not found in the present study of Danish white subjects. The effect of this variant on obesity in Danish subjects is contentious and further study is needed.
Similar content being viewed by others
Introduction
Type 2 diabetes is a rapidly growing public health problem with a tremendous impact on morbidity and mortality worldwide. Although the epidemic nature of the disease may be attributable mainly to environmental factors leading to obesity, genetic factors also predispose to the disease. A characteristic feature of type 2 diabetes is insulin resistance in skeletal muscle, liver and adipose tissue, resulting from abnormalities in peripheral insulin signalling through the insulin receptor and a subsequent intracellular signalling network involving a complex cascade of events [1, 2]. Although it is well accepted that insulin resistance is also under genetic control and that this predisposition modulates diabetes susceptibility, the genes involved are still mostly unknown.
The ectoenzyme nucleotide pyrophosphatase phosphodiesterase, also known as plasma cell membrane glycoprotein 1 (PC-1) inhibits insulin signalling by direct interaction with the insulin receptor α subunit [3, 4]. PC-1 reduces both insulin receptor function and subsequent downstream signalling, and has a wide range of tissue expression that includes skeletal muscle and the liver [5, 6]. Furthermore, protein levels of PC-1 are increased in insulin-resistant subjects and correlate with whole-body insulin resistance [7]. Overexpression of human PC-1 in the liver of mice induces insulin resistance and glucose intolerance [8], suggesting that this protein influences insulin sensitivity and thereby may affect the risk of developing type 2 diabetes.
The involvement of PC-1 in insulin-resistant states is further supported by recent studies revealing evidence for linkage between the chromosomal region 6q22–q23 of the gene encoding PC-1 (ENPP1) and insulin resistance [9], obesity [10, 11] and type 2 diabetes [12–15]. Furthermore, the minor Q allele of a K121Q variant (rs1044498) of ENPP1 has been shown to influence PC-1 protein function by inhibiting insulin receptor function and insulin signalling more effectively than the major K allele [16, 17]. Likewise, overexpression of the ENPP1 Q allele in mouse muscle and liver tissues causes insulin resistance and glucose intolerance in vivo [18].
In contrast, outcomes of human studies of the K121Q variant have been conflicting. Hence, initial findings indicating association of the Q allele with insulin resistance [16, 19, 20] were not confirmed in all subsequent studies [21, 22]. Some studies have been able to relate the Q allele to type 2 diabetes [23, 24] whereas others failed to do so [19, 21, 25–27]. Likewise, a large intervention study failed to show evidence of an impact of the K121Q variant on the conversion from IGT to overt type 2 diabetes or weight change during 3 years of follow-up [28], whereas another prospective investigation found the Q allele to be associated with deterioration in an atherogenic risk profile as well as an earlier onset of type 2 diabetes [20]. Recently, evidence for an association of the K121Q variant and a risk haplotype defined by three single-nucleotide polymorphisms (SNPs) in ENPP1, including the K121Q variant, with different subtypes of obesity and type 2 diabetes was demonstrated in a sample of about 6,000 subjects of French and Austrian Caucasian origin [29].
Given these ambiguous findings, the objective of the present study was to investigate in a relatively large-scale setting the ENPP1 K121Q variant in relation to type 2 diabetes and associated metabolic quantitative traits in samples of Danish white subjects.
Subjects and methods
Subjects
The case–control studies of overweight and obesity were performed in the Inter99 cohort, which is a population-based, randomised, non-pharmacological intervention study for the prevention of ischaemic heart disease conducted at the Research Centre for Prevention and Health in Glostrup, Copenhagen (ClinicalTrials.gov; identifier: NCT00289237) [30]. Patients treated for type 2 diabetes (n=98) were excluded. The subjects (n=5,863) were divided into three classes according to BMI: (1) lean subjects with BMI<25.0 kg/m2 (n=2,582); (2) overweight subjects with BMI in the range 25.0–29.9 kg/m2 (n=2,312); and (3) obese subjects with BMI ≥30.0 kg/m2 (n=969).
The case–control study of diabetes included 1,386 (836 men, 550 women) unrelated patients with type 2 diabetes. Diabetes was diagnosed according to the World Health Organization 1999 criteria [31]. Of the patients with type 2 diabetes, 1,042 were recruited at the outpatient clinic at Steno Diabetes Center and 344 (98 with known type 2 diabetes and 246 with screen-detected type 2 diabetes) were from the Inter99 cohort [30]. The phenotypic characteristics of the cases (mean±SD) were: age at entry to the examination, 56.9±10.5 years; age at clinical diagnosis, 51.9±10.6 years; BMI, 29.7±5.3 kg/m2; and HbA1C, 7.8±1.7%. At recruitment, patients with one or more of the following characteristics were excluded: (1) diabetes due to known chronic pancreatitis, haemochromatosis, severe insulin resistance, MODY, or maternally inherited diabetes and deafness; (2) a family history of first-degree relatives with type 1 diabetes; (3) a requirement for insulin within the first year after the diagnosis of diabetes; or (4) a fasting serum C-peptide level ≤150 pmol/l. Among the patients with type 2 diabetes, 130 (71 men, 59 women) were aged 40 years or younger at clinical diagnosis; this group was termed ‘early-onset type 2 diabetes’. The phenotypic characteristics of this subgroup (mean±SD) were: age at entry to the examination, 44.9±9.2 years; age at clinical diagnosis, 34.5±6.0 years; BMI, 30.7±5.9 kg/m2; and HbA1C, 8.2±1.8%. The control group comprised 4,440 unrelated subjects from the Inter99 cohort recruited in Copenhagen County and 330 unrelated subjects recruited at random from Copenhagen County (mean±SD age 46.3±8.8 years, BMI 25.6±4.0 kg/m2). All control subjects underwent a standard 75 g OGTT and only subjects who had fasting normoglycaemia and were normal glucose tolerant were included as control subjects in the study.
The genotype–quantitative trait study was carried out in the Inter99 cohort [30], including subjects with normoglycaemia and NGT (n=4,440), impaired fasting glycaemia (IFG) (n=495), impaired glucose tolerance (IGT) (n=682) and screen-detected and treatment-naive type 2 diabetes (n=246).
All study participants were Danish white subjects by self-report. Informed written consent was obtained from all study participants. The studies were conducted in accordance with the Declaration of Helsinki II and were approved by the local Ethical Committee of Copenhagen.
Biochemical and anthropometric measures
Blood samples were taken after a 12-h overnight fast. An OGTT was performed in all non-diabetic subjects. The HbA1C (normal range 4.1–6.4%), plasma glucose and serum insulin concentrations were measured as described previously [32].
Genotyping
Genomic DNA was purified from human leucocytes obtained from whole blood using standard methods [33]. Genotyping of the ENPP1 K121Q polymorphism (rs1044498) was performed by chip-based matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (DNA MassARRAY; Sequenom, San Diego, CA, USA) of PCR-generated primer extension products, as described previously [34]. The polymorphism was genotyped with a success rate of 98.1%. One hundred and eighty samples were genotyped in duplicate and no mismatches were found. The genotypes were in Hardy–Weinberg equilibrium (p=0.14).
Statistical analysis
In the case–control studies of type 2 diabetes and obesity, Fisher’s exact test was used to examine differences in allele frequencies between cases and control subjects and logistic regression was used to examine differences in genotype distributions with adjustment for sex, age and BMI. A general linear model was used for testing quantitative traits in relation to genotype. Genotype and sex were entered in the model as fixed factors, and age and BMI as covariates. A p-value of less than 0.05 was considered significant. Logistic regression and analyses on quantitative traits were performed using Statistical Package for Social Science software (version 13.0; SPSS, Chicago, IL, USA). A meta-analysis of the present and published studies was performed using RGui version 2.2.1 (available at http://www.r-project.org). Homogeneity between studies and the meta-analysis using the Mantel–Haenszel method was tested with a generalised linear model. Publication bias was examined by unweighted regression analysis of funnel plot asymmetry [35] and qq plots of standardized effect-size estimates [36].
Results
We performed separate case–control studies of the ENPP1 K121Q polymorphism (rs1044498) with diabetes, overweight and obesity.
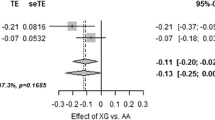
In the case–control study of type 2 diabetes we found no differences in allele frequencies and genotype distributions between 1,386 unrelated patients with type 2 diabetes and 4,770 glucose-tolerant subjects (Table 1). Likewise, assuming either a dominant or a recessive model for the penetrance of the minor Q allele, no significant differences were observed between groups (data not shown). Repeated analysis using an age-matched control subgroup (mean age±SD, 56.1±4.8 years, n=1,588) did not change the results (data not shown). Furthermore, no association of the K121Q genotype with early-onset type 2 diabetes (clinical diagnosis at 40 years or younger) was detected (data not shown). To test more specifically for an association with obesity-related type 2 diabetes, we then included only patients with type 2 diabetes with a BMI above 30 kg/m2 (n=570) and compared allele frequency and genotype distribution with those of a lean control subgroup (BMI<25 kg/m2; n=2,352). No association with obesity-related type 2 diabetes was observed (data not shown). We performed a meta-analysis of the present study and all previously published case–control studies of type 2 diabetes [16, 19, 20, 23–27, 29] except [37], for which data were not accessible, including a total of 6,466 patients with type 2 diabetes and 11,220 control subjects (Fig. 1). No heterogeneity between studies was detected (p=0.11) and no evidence for publication bias was found using both unweighted regression (p=0.51) [35] and qq plots of standardized effect-size estimates (all dots in the qq plot were within the 95% CI) [36]. We observed evidence of association of the ENPP1 codon 121 Q allele with type 2 diabetes with an odds ratio (OR) per Q allele of 1.17 (95% CI 1.10–1.25, p=1×10−6).
Estimated odds ratio (95% CI) of type 2 diabetes when carrying the ENPP1 codon 121 Q allele (KQ and QQ vs KK genotypes) in a meta-analysis of the present and published case–control studies [16, 19, 20, 23–27, 29] except [37], for which data were not accessible. Numbers in round brackets designate the numbers of type 2 diabetic patients and control subjects; those in square brackets indicate the reference number. Test of homogeneity between studies, p=0.11. Odds ratio for the risk of type 2 diabetes per Q allele, 1.17 (95% CI 1.10–1.25, p=1×10−6)
In the case–control study of overweight and obesity, we compared allele frequencies and genotype distributions between different strata of BMI among 5,863 middle-aged subjects of a population-based sample. In a recessive model, we found an uncorrected, statistically significant association with being overweight in QQ allele carriers when comparing overweight subjects (BMI≥25 kg/m2) with normal-weight subjects (BMI<25 kg/m2) (OR 1.63 [95% CI 1.09–2.46]; p=0.015). No significant effect of genotype was, however, found when applying a dominant model or comparing allele frequencies (Table 2). When comparing obese subjects (BMI≥30 kg/m2) with either normal-weight subjects (BMI<25 kg/m2) or normal-weight and overweight subjects (BMI<30 kg/m2), no significant associations were found (Table 2). Similar results were obtained when adjusting for the effects of age and sex (data not shown).
To try to replicate previous findings of an influence of the ENPP1 gene on more extreme forms of obesity [29, 38], we compared the K121Q genotypes of the subjects above the 90th percentile of the BMI distribution (n=586, mean±SD BMI, 35.7±3.8 kg/m2) with the genotypes of subjects with BMI between the 10th and 20th percentiles (n=583, mean±SD BMI, 21.9±0.4 kg/m2). No differences in allele frequencies or genotype distributions were found between these groups (p=0.6 for the codominant model; p=0.4 for the dominant model for the Q allele; p=0.6 for the recessive model for the Q allele).
In the genotype–quantitative trait study of a population-based sample of middle-aged participants including subjects with NGT, IFG, IGT and screen-detected type 2 diabetes, we evaluated anthropometric measures, fasting and post-OGTT levels of plasma glucose, serum insulin and serum C-peptide and measures of insulin resistance. We found no significant effect of the K121Q genotype on these variables (Table 3).
Apart from a probably random finding of a difference in serum C-peptide level at 30 min (p=0.04 for the codominant model), no significant associations were observed in the subgroup of NGT subjects (n=4,440) (data not shown).
Discussion
Previous studies indicate that PC-1 modulates insulin signalling through inhibition of the insulin receptor, presumably by a direct interaction with the α subunit, and in vitro data and studies in rodents further suggest that this effect is modified by the K121Q polymorphism of ENPP1 [3, 4, 17]. It has also been hypothesised that the increased diabetes susceptibility that might be associated with the K121Q polymorphism derives from the effect on insulin sensitivity. However, studies of the clinical impact of the polymorphism in humans have generated conflicting results.
In the present study we found a putative association of the ENPP1 codon 121 QQ genotype with being overweight, defined as BMI≥25 kg/m2. It is possible that this is a true association, but we cannot rule out a spurious positive association due to multiple testing. In favour of the first position is the fact that previous studies support a recessive model for the Q allele on measures of obesity [26, 29], yet further analyses showed that the nominal association in the present study was only evident at a BMI cut-off between 24.0 and 25.5 kg/m2. In addition, analysis of BMI as a quantitative trait revealed no association with K121Q genotype (Table 3). No evidence of an impact of the variant on measures of insulin sensitivity indices [39–41] was observed. In fact, the impact of the gene variant on insulin resistance has been ambiguous, yielding association in some but not all populations studied [16, 19–22, 42]. Finally, we were unable to relate the K121Q variant to the presence of type 2 diabetes or associated quantitative metabolic traits in the Danish sample.
To date, nine case–control studies [19–21, 23–27, 29] testing association of the K121Q variant with type 2 diabetes have been published. No definitive role for the involvement of the polymorphism in the pathogenesis of type 2 diabetes has been established on the basis of these investigations. Thus, in most studies of the K121Q variant in Caucasian subjects, all with a limited number of cases and control subjects, no relationship with type 2 diabetes was found [19–21, 26, 27], whereas evidence of association has been obtained in four populations published in two reports [23, 29]. Yet the present updated meta-analysis of the current and published case–control studies yielded evidence of an association with type 2 diabetes with an OR of 1.17 per Q allele. Interestingly, the meta-analysis supported an additive genetic model as we were able to reject both the dominant (p=0.004) and the recessive model (p=0.001) in favour of the full model. Still it should be pointed out that meta-analysis, which in principle might be expected to provide conclusive answers, may, in the area of genetic epidemiology, be compromised by heterogeneity of ethnicity and outcome phenotypes besides publication and ascertainment bias. In the present meta-analysis we failed to show evidence for publication bias using two different approaches [35, 36]; however, both methods are weakened by lack of power due to the relatively small number of studies in the meta-analysis.
Recently, a study comprising two larger samples of 1,308 French and 1,261 Austrian Caucasians, demonstrated association of a three-allele risk ENPP1 haplotype including the K121Q variant with type 2 diabetes, with an OR of 1.56 in the pooled analysis of 1,255 patients with type 2 diabetes and 1,314 control subjects [29]. Additionally, this study revealed highly significant associations between the risk haplotype as well as the K121Q variant analysed separately and severe forms of obesity (696 morbidly obese [BMI>40 kg/m2] and 717 moderately obese [BMI 30–40 kg/m2] subjects), including childhood obesity (529 subjects with BMI above the 97th percentile for age and sex) [29]. Despite our large sample size, we failed to replicate this finding convincingly when analysing the K121Q variant in multiple BMI strata. However, importantly, the designs of the French study and the present protocol differ considerably, especially regarding the sampling of patients. Meyre et al. recruited cases according to the severity of obesity, focusing on the most severe forms of obesity, whereas our investigation was performed using a population-based sampling design, thereby concentrating on more common forms of obesity. Consequently, our ability to obtain evidence for an effect of K121Q genotype on morbid obesity is limited. Besides, we did not examine the full three-SNP haplotype, a fact that confines comparisons between the two studies somewhat, since it is possible, as indicated by the French study [29], that more than one variant in ENPP1 modulates the risk of type 2 diabetes and obesity. A few other smaller (of the order of 400 to 1,100 participants) and probably statistically underpowered studies found either no association [19, 22, 37, 42], association of the rare Q allele [26] or association of the common K allele with obesity [38].
The reason for the apparent discrepancies between the studies, including ours, that have evaluated the pathogenic impact of the K121Q variant is far from obvious. It is possible that the susceptibility induced by the polymorphism is modulated by interactions with other ethnic-specific genetic or environmental factors and that the phenotypic expression of the variant will therefore be different in various ethnic populations. In this regard it is emphasised that the frequency of the K121Q polymorphism varies considerably between different ethnic groups [24, 38, 42, 43]. It is also recognised that many case–control studies of multifactorial diseases with inheritance as complex as that of type 2 diabetes and obesity are often statistically underpowered in order to yield conclusive results, and random spurious positive associations due to multiple testing often cannot be excluded. We believe that studies that are adequately powered, as was the study described in the present report, are required; among the published association studies of the K121Q variant, ours is the largest so far. Thus, on the basis of the previous reports of the presumed effect of the K121Q genotype [20, 29] we estimate the statistical power at a significance level of 0.05 in the present study to detect an association with type 2 diabetes and obesity (depending on BMI strata) to be 85–98% and 90–99%, respectively.
On the basis of the present study, performed in a relatively large study sample of ethnically homogeneous people, we conclude that the ENPP1 K121Q polymorphism does not seem to influence the risk of insulin resistance or type 2 diabetes in Danish white subjects; however, a meta-analysis of the present and published studies demonstrates evidence of association with type 2 diabetes. A possible effect of the ENPP1 K121Q variant on the risk of obesity needs to be confirmed in large population-based studies and samples enriched for severe obesity.
Abbreviations
- HOMA-IR:
-
homeostasis model assessment of insulin resistance
- ISI:
-
insulin sensitivity index
- MAF:
-
minor allele frequency
- OR:
-
odds ratio
- PC-1:
-
plasma cell membrane glycoprotein 1
- SNP:
-
single nucleotide polymorphism
References
Rhodes CJ, White MF (2002) Molecular insights into insulin action and secretion. Eur J Clin Invest 32(Suppl 3):3–13
DeFronzo RA, Bonadonna RC, Ferrannini E (1992) Pathogenesis of NIDDM: a balanced overview. Diabetes Care 15:318–368
Maddux BA, Sbraccia P, Kumakura S et al (1995) Membrane glycoprotein PC-1 and insulin resistance in non-insulin-dependent diabetes mellitus. Nature 373:448–451
Maddux BA, Goldfine ID (2000) Membrane glycoprotein PC-1 inhibition of insulin receptor function occurs via direct interaction with the receptor α-subunit. Diabetes 49:13–19
Harahap AR, Goding JW (1988) Distribution of the murine plasma cell antigen PC-1 in non-lymphoid tissues. J Immunol 141:2317–2320
Kumakura S, Maddux BA, Sung CK (1998) Overexpression of membrane glycoprotein PC-1 can influence insulin action at a post-receptor site. J Cell Biochem 68:366–377
Frittitta L, Youngren J, Vigneri R, Maddux BA, Trischitta V, Goldfine ID (1996) PC-1 content in skeletal muscle of non-obese, non-diabetic subjects: relationship to insulin receptor tyrosine kinase and whole body insulin sensitivity. Diabetologia 39:1190–1195
Dong H, Maddux BA, Altomonte J et al (2005) Increased hepatic levels of the insulin receptor inhibitor, PC-1/NPP1, induce insulin resistance and glucose intolerance. Diabetes 54:367–372
Duggirala R, Blangero J, Almasy L et al (2001) A major locus for fasting insulin concentrations and insulin resistance on chromosome 6q with strong pleiotropic effects on obesity-related phenotypes in nondiabetic Mexican Americans. Am J Hum Genet 68:1149–1164
Meyre D, Lecoeur C, Delplanque J et al (2004) A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31–q23.2. Diabetes 53:803–811
Atwood LD, Heard-Costa NL, Cupples LA, Jaquish CE, Wilson PW, D’Agostino RB (2002) Genomewide linkage analysis of body mass index across 28 years of the Framingham Heart Study. Am J Hum Genet 71:1044–1050
Ehm MG, Karnoub MC, Sakul H et al (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881
Ghosh S, Watanabe RM, Valle TT et al (2000) The Finland–United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study I An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185
Demenais F, Kanninen T, Lindgren CM et al (2003) A meta-analysis of four European genome screens (GIFT Consortium) shows evidence for a novel region on chromosome 17p11.2–q22 linked to type 2 diabetes. Hum Mol Genet 12:1865–1873
Xiang K, Wang Y, Zheng T et al (2004) Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21–q23 and chromosome 1q21–q24. Diabetes 53:228–234
Pizzuti A, Frittitta L, Argiolas A et al (1999) A polymorphism (K121Q) of the human glycoprotein PC-1 gene coding region is strongly associated with insulin resistance. Diabetes 48:1881–1884
Costanzo BV, Trischitta V, Di Paola R et al (2001) The Q allele variant (GLN121) of membrane glycoprotein PC-1 interacts with the insulin receptor and inhibits insulin signaling more effectively than the common K allele variant (LYS121). Diabetes 50:831–836
Maddux BA, Chang YN, Accili D, McGuinness OP, Youngren JF, Goldfine ID (2006) Overexpression of the insulin receptor inhibitor PC-1/ENPP1 induces insulin resistance and hyperglycemia. Am J Physiol Endocrinol Metab 290:E746–E749
Gu HF, Almgren P, Lindholm E et al (2000) Association between the human glycoprotein PC-1 gene and elevated glucose and insulin levels in a paired-sibling analysis. Diabetes 49:1601–1603
Bacci S, Ludovico O, Prudente S et al (2005) The K121Q polymorphism of the ENPP1/PC-1 gene is associated with insulin resistance/atherogenic phenotypes, including earlier onset of type 2 diabetes and myocardial infarction. Diabetes 54:3021–3025
Rasmussen SK, Urhammer SA, Pizzuti A et al (2000) The K121Q variant of the human PC-1 gene is not associated with insulin resistance or type 2 diabetes among Danish Caucasians. Diabetes 49:1608–1611
Gonzalez FJ, Martinez-Larrad MT, Fernandez P, Kubaszek A, Laakso M, Serrano R (2003) K121Q PC-1 gene polymorphism is not associated with insulin resistance in a Spanish population. Obes Res 11:603–605
Abate N, Chandalia M, Satija P et al (2005) ENPP1/PC-1 K121Q polymorphism and genetic susceptibility to type 2 diabetes. Diabetes 54:1207–1213
Hamaguchi K, Terao H, Kusuda Y et al (2004) The PC-1 Q121 allele is exceptionally prevalent in the Dominican Republic and is associated with type 2 diabetes. J Clin Endocrinol Metab 89:1359–1364
Hegele RA, Harris SB, Zinman B, Hanley AJ, Cao H (2001) Absence of association of type 2 diabetes with CAPN10 and PC-1 polymorphisms in Oji-Cree. Diabetes Care 24:1498–1499
Barroso I, Luan J, Middelberg RP et al (2003) Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol 1:e20
Keshavarz P, Inoue H, Sakamoto Y et al (2006) No evidence for association of the ENPP1 (PC-1) K121Q variant with risk of type 2 diabetes in a Japanese population. J Hum Genet 51:559–566
Laukkanen O, Pihlajamäki J, Lindström J et al (2004) Common polymorphisms in the genes regulating the early insulin signalling pathway: effects on weight change and the conversion from impaired glucose tolerance to type 2 diabetes. Diabetologia 47:871–877
Meyre D, Bouatia-Naji N, Tounian A et al (2005) Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet 37:863–867
Jørgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glumer C, Pisinger C (2003) A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99. Eur J Cardiovasc Prev Rehabil 10:377–386
World Health Organization Study Group (1999) Part 1: Diagnosis and classification of diabetes mellitus. Technical Report Series, no. WHO/NCD/NCS/99.2. World Health Organization, Geneva
Andersen G, Overgaard J, Albrechtsen A et al (2006) Studies of the association of the GNB3 825C>T polymorphism with components of the metabolic syndrome in white Danes. Diabetologia 49:75–82
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Buetow KH, Edmonson M, MacDonald R et al (2001) High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci USA 98:581–584
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Wang MC, Bushman BJ (1998) Using the normal quantile plot to explore meta-analytic data sets. Psychol Methods 3:46–54
Kubaszek A, Markkanen A, Eriksson JG et al (2004) The association of the K121Q polymorphism of the plasma cell glycoprotein-1 gene with type 2 diabetes and hypertension depends on size at birth. J Clin Endocrinol Metab 89:2044–2047
Matsuoka N, Patki A, Tiwari HK et al (2006) Association of K121Q polymorphism in ENPP1 (PC-1) with BMI in Caucasian and African-American adults. Int J Obes (Lond) 30:233–237
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419
Matsuda M, DeFronzo RA (1999) Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22:1462–1470
Belfiore F, Iannello S, Camuto M, Fagone S, Cavaleri A (2001) Insulin sensitivity of blood glucose versus insulin sensitivity of blood free fatty acids in normal, obese, and obese-diabetic subjects. Metabolism 50:573–582
Kubaszek A, Pihlajamaki J, Karhapaa, P, Vauhkonen I, Laakso M (2003) The K121Q polymorphism of the PC-1 gene is associated with insulin resistance but not with dyslipidemia. Diabetes Care 26:464–467
Morrison JA, Gruppo R, Glueck CJ et al (2004) Population-specific alleles: the polymorphism (K121Q) of the human glycoprotein PC-1 gene is strongly associated with race but not with insulin resistance in black and white children. Metabolism 53:465–468
Acknowledgements
The authors wish to thank A. Forman, I.-L. Wantzin and M. Stendal for technical assistance, G. Lademann for secretarial support and B. Carstensen for statistical support. The study was supported by grants from the European Union (EUGENE2, grant no. LSHM-CT-2004-512013), the Danish Medical Research Council, the Danish Diabetes Association, the Velux Foundation, the Sehested Hansen Foundation and the Danish Heart Association.
Duality of interest
K.B.-J. holds stock in and has received research support from Novo Nordisk A/S.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grarup, N., Urhammer, S.A., Ek, J. et al. Studies of the relationship between the ENPP1 K121Q polymorphism and type 2 diabetes, insulin resistance and obesity in 7,333 Danish white subjects. Diabetologia 49, 2097–2104 (2006). https://doi.org/10.1007/s00125-006-0353-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-006-0353-x