Abstract
Aims/hypothesis
AMP-activated protein kinase (AMPK) regulates metabolic adaptations in skeletal muscle. The aim of this study was to investigate whether AMPK modulates the expression of skeletal muscle genes that have been implicated in lipid and glucose metabolism under fed or fasting conditions.
Methods
Two genetically modified animal models were used: AMPK γ3 subunit knockout mice (Prkag3 −/−) and skeletal muscle-specific transgenic mice (Tg-Prkag3 225Q) that express a mutant (R225Q) γ3 subunit. Levels of mRNA transcripts of genes involved in lipid and glucose metabolism in white gastrocnemius muscles of these mice (under fed or 16-h fasting conditions) were assessed by quantitative real-time PCR.
Results
Wild-type mice displayed a coordinated increase in the transcription of skeletal muscle genes encoding proteins involved in lipid/oxidative metabolism (lipoprotein lipase, fatty acid transporter, carnitine palmitoyl transferase-1 and citrate synthase) and glucose metabolism (glycogen synthase and lactate dehydrogenase) in response to fasting. In contrast, these fasting-induced responses were impaired in Prkag3 −/− mice. The transcription of genes involved in lipid and oxidative metabolism was increased in the skeletal muscle of Tg-Prkag3 225Q mice compared with that in wild-type mice. Moreover, the expression of the genes encoding hexokinase II and 6-phosphofrucktokinase was decreased in Tg-Prkag3 225Q mice after fasting.
Conclusions/interpretation
AMPK is involved in the coordinated transcription of genes critical for lipid and glucose metabolism in white glycolytic skeletal muscle.
Similar content being viewed by others
Introduction
Living organisms are subjected to intermittent food supply, and the ability to cope with such changes is vital for survival. In Saccharomyces cerevisiae, glucose depletion induces a shift from the anaerobic metabolism of glucose to the aerobic metabolism of alternative carbon sources [1–3]. This shift in substrate use is modulated at the transcriptional level, at least in part, by sucrose non-fermenting protein kinase 1 (SNF1) [1–3]. In mammalian skeletal muscle, the flexibility of carbon source utilisation is conserved. Under fed conditions, glucose is the main fuel source in skeletal muscle. However, during fasting, fuel use shifts to lipid metabolism, concomitantly with an increase in the expression of genes encoding proteins that regulate lipid metabolism, such as lipoprotein lipase (LPL), carnitine palmitoyl transferase-1 (CPT-1) and uncoupling protein 3 (UCP3) [4–6]. Thus, metabolic flexibility of skeletal muscle plays a central role in whole-body energy homeostasis and the pathogenesis of metabolic diseases such as type 2 diabetes.
AMP-activated protein kinase (AMPK), the mammalian homologue of SNF1 protein kinase, affects metabolism by post-translational modulation via allosteric interaction or covalent modification [1, 3, 7]. AMPK induces skeletal muscle glucose uptake by promoting translocation of GLUT4 to the cell surface [8] and lipid metabolism via inactivation of acetyl-CoA carboxylase (ACC) [9]. Treatment of rodents with 5-aminoimidazole-4-carboxamide-1-β-4-ribofuranoside (AICAR; a pharmacological activator of AMPK) increases the expression of genes for proteins involved in substrate and energy metabolism, such as GLUT4, hexokinase II (HK2) and mitochondrial proteins (including cytochrome c [CYCS], δ-aminolevulinic acid synthase and UCP3) in skeletal muscle [10–13], indicating that AMPK and SNF1 protein kinase may play similar roles in transcriptional regulation. Although fasting induces transcriptional adaptations in skeletal muscle, the direct role of AMPK in the modulation of genes involved in glucose and alternative carbon fuel metabolism is unclear.
The α, δ and γ isoforms of the peroxisome proliferator-activated receptor (PPAR) are implicated in the transcriptional regulation of genes essential for lipid homeostasis [14, 15]. PPARα mediates the metabolic response to fasting by increasing the transcription of genes essential for fatty acid oxidation in the liver [16, 17] and heart [16]. PPARγ regulates adipocyte differentiation and lipid storage by modulating the expression of genes involved in fatty acid oxidation [14, 18], and regulates whole-body glucose uptake by mediating insulin action in skeletal muscle [19]. The function of PPARδ remains largely elusive; however, overexpression of the gene for an activated PPARδ in mouse adipose tissue induced expression of genes required for fatty acid oxidation and energy dissipation, improved lipid profiles and reduced adiposity [20]. Although PPARs and AMPK regulate diverse metabolic responses, evidence for a role of AMPK in modulating the transcription of PPARs is lacking.
Mammalian AMPK is a structurally conserved heterotrimer consisting of catalytic α subunits and regulatory β and γ subunits [1, 3, 7]. The γ subunit includes γ1, γ2 and γ3 isoforms, with γ3 being the predominant isoform in glycolytic skeletal muscle [21], a fibre type that depends upon the anaerobic metabolism of glucose when there is a high demand for energy. In skeletal muscle from pigs carrying a naturally occurring mutation of γ3 (R225Q), glycogen content [22, 23] and the activities of citrate synthase (CS) and hydroxyacyl-CoA dehydrogenase are increased [24, 25], suggesting that the γ3 subunit can regulate metabolic properties of skeletal muscle.
The aim of this study was to investigate the role of the γ3 subunit of AMPK in the transcriptional modulation of genes implicated in glucose and lipid metabolism, as well as those for the PPARs isoforms, under fed and fasting conditions in skeletal muscle. Two genetically modified animal models were utilised: AMPK γ3 subunit (Prkag3) knockout mice (Prkag3 −/−) and skeletal muscle-specific transgenic mice (Tg-Prkag3 225Q) harbouring a mutant (R225Q) γ3 subunit [26]. Genetic ablation of the γ3 subunit abolishes AICAR-induced glucose uptake in skeletal muscle in vitro, suggesting a critical role for the subunit in AMPK function [26]. Furthermore, under fasting conditions, AMPK activity in skeletal muscle of Prkag3 −/− mice is lower than in wild-type mice [27]. In vitro studies provide evidence that R225Q is an activating mutation, such that AMPK activity is higher in heterotrimeric complexes containing the mutant vs wild-type γ3 subunit AMPK [26]. We have also reported previously that phosphorylation of ACC is increased in skeletal muscle of Tg-Prkag3 225Q mice [26]. Here, we provide evidence that AMPK is involved in modulating key genes regulating lipid and glucose metabolism in skeletal muscle.
Materials and methods
AMPK γ3 knockout and γ3225Q transgenic mice
The AMPK γ3 gene (Prakg3) knockout mice (Prkag3 −/−) and skeletal muscle-specific transgenic mice that express a mutant γ3 subunit (Tg-Prkag3 225Q) have been previously described [26]. AMPK Prkag3 −/− mice were created by conventional gene targeting techniques. The Tg-Prkag3 225Q mice express the mutant γ3225Q subunit under the control of mouse myosin light chain promoter and enhancer.
Mice were maintained in a light–dark cycle of 12 h each, and were cared for in accordance with regulations for the protection of laboratory animals. The regional animal ethics committee approved all experimental procedures. Mice (10–12 weeks old) were given free access to water and standard rodent chow. When examining the effects of fasting, food was removed 16 h prior to study. The white portion of the gastrocnemius skeletal muscle was removed from anaesthetised mice, cleaned of fat and blood, and quickly frozen in liquid nitrogen.
RNA purification and cDNA synthesis
Skeletal muscle (50 mg) was homogenised in 1 ml of Trizol reagent (Sigma, St Louis, MO, USA) and total RNA was purified according to the manufacturer’s recommendations. Purified RNA (3 μg) was then treated with 6 U of DNAse I, using the DNA-free kit (Ambion, Huntingdon, UK) according to the manufacturer’s protocol. DNAse-treated RNA was used as a template for cDNA synthesis with oligo(dT) primers using the SuperScript First Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). A reaction without reverse transcriptase was performed for each sample as a control.
Quantitative PCR
Levels of gene expression were quantified using real-time PCR with the ABI PRISM 7000 Sequence Detector System and fluorescence-based SYBR Green technology (Applied Biosystems, Warrington, UK). PCR was performed in a final volume of 25 μl. The reaction consisted of diluted cDNA sample, SYBR Green PCR Master Mix (Applied Biosystems), primers optimised for each target transcript, and nuclease-free water. Data were analysed using the standard curve method. The relative quantities of target transcripts were calculated from duplicate samples after normalisation of the data against the housekeeping gene (encoding acidic ribosomal phosphoprotein PO [ARBP]). Primers were selected by using Primer Express computer software (Applied Biosystems). Transcript sequences were obtained from (1) the ENSEMBL database (available at http://www.ensembl.org/index.html, last accessed in August 2005) – CPT-1 (Cpt1, ENSMUST00000052315), CD36 (Cd36, ENSMUST00000003024), CS (Cs, ENSMUST00000005826), GLUT4 (Slc2a4, ENSMUST00000018710), glycogen synthase (Gys, ENSMUST00000003964), short-chain 3-hydroxyacyl-CoA dehydrogenase (Hadhsc, ENSMUST00000029610), LPL (Lpl, ENSMUST00000015712), muscle 6-phosphofructokinase (Pfkm, ENSMUST00000043950), pyruvate dehydrogenase kinase 4 (Pdk4, ENSMUST00000019721), UCP3 (Ucp3, ENSMUST00000032958); and (2) the National Center for Biotechnology Information (NCBI) Genbank database (available at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide) – ARBP (Arbp, BC003833), CYCS (Cycs, NM007808), HK2 (Hk2, Y11666), lactate dehydrogenase 2 (LDH2) B chain (Ldh2, NM008492), PPARα (Ppara, NM011144), PPARδ (Ppard, NM011145) and PPARγ (Pparg, NM011146).
Western blot analysis
White gastrocnemius muscle (30 mg) was pulverised in microcentrifuge tubes over liquid nitrogen. Powdered muscle was homogenised in 400 μl of ice-cold lysis buffer (250 mmol/l sucrose, 1 mmol/l EDTA, 20 mmol/l Tris-HCl, pH 7.5) by an electrical homogeniser. The homogenate was centrifuged (800 g for 15 min at 4°C), and the pellet was homogenised and centrifuged (800 g for 15 min at 4°C). The supernatant fractions were combined and centrifuged (190,000 g for 1 h at 4°C). The supernatant fraction (cytoplasmic fraction) was collected, and the pellet of crude membranes was dissolved in lysis buffer. The total protein content of the cytoplasmic and membrane fractions was determined using a commercially available kit based on the Bradford method (Bio-Rad, Hercules, CA). The cytoplasmic fraction was used for determination of GYS and HK2 content, while the membrane fraction was used for analysis of CD36, GLUT4 (now known as SLC2A4) and UCP3 content. Proteins were solubilised in Laemmli sample buffer, separated by SDS-PAGE and transferred onto Immobilon-P membranes (Millipore, Bedford, MA, USA). Membranes were blocked with 7.5% fat-free milk in Tris-buffered saline containing 0.02% Tween-20 (TBST) and then probed with the specified primary antibodies. The GLUT4 antibody was from Biogenesis (Poole, UK). GYS and HK2 antibodies were kind gifts from O. Pedersen (Steno Memorial Hospital, Gentofte, Denmark). CD36 and UCP3 antibodies were from Cayman Chemical (Ann Arbor, MI, USA) and Santa Cruz Biotechnology (Santa Cruz, CA, USA), respectively. Membranes were washed with TBST and incubated with an appropriate secondary antibody. Proteins were visualised by enhanced chemiluminescence and quantified by densitometry.
Glycogen and triglyceride analyses
For glycogen determinations, skeletal muscle (4–10 mg) was homogenised in 0.5 ml of 1 mol/l HCl (100°C for 1 h). Glycogen was measured fluorometrically [28]. For triglyceride determinations, skeletal muscle (15–20 mg) was homogenised with 0.3 ml of heptane–isopropanol–Tween mixture (3:2:0.01, by volume) and centrifuged (1,500 g for 15 min at 4°C). The upper phase (containing extracted triglycerides) was collected and evaporated with vacuum centrifuge. The triglyceride content was measured in duplicate samples using a glycerol-blanked kit (Roche, Mannheim, Germany) and Seronorm lipid (SERO, Billingstad, Norway) standards.
Statistical analyses
Differences among groups were determined by two-way ANOVA followed by Fisher’s least significant differences post hoc analysis. A p value of less than 0.05 was considered statistically significant.
Results
Genes implicated in lipid metabolism
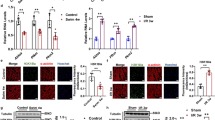
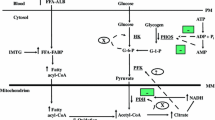
There are several sites of regulation in lipid metabolism in skeletal muscle. The entry of fatty acid into the sarcoplasm is regulated by selective transport of NEFAs across the sarcoplasm (Fig. 1). Fatty acids are transported systemically as triglycerides and are liberated by LPL. Under fasting conditions, Lpl expression in wild-type mice was augmented (81%, p<0.05) (Fig. 2a). This effect was impaired in Prkag3 −/− mice (Fig. 2a).
Lipid and glucose metabolism of skeletal muscle. Schematic representation of the pathways of glucose and lipid metabolism in skeletal muscle, with acetyl-CoA as the major conjunctive metabolite of glycolysis and lipid oxidation. FA−, fatty acid anion; fructose 1,6-bi-P, fructose 1,6-bisphosphate; glucose 6-P, glucose 6-phosphate; PDH, pyruvate dehyrogenase; TG, triglyceride
Relative expression of genes regulating lipid metabolism. The expression of Lpl (a), Cd36 (b), Cpt1 (c), Hadhsc (d), Cs (e), Cycs (f) and Ucp3 (g), which encode proteins that regulate lipid metabolism, in white glycolytic gastrocnemius muscle of fed (white bars) and fasted (black bars) wild-type (WT), Tg-Prkag3 225Q and Prkag3 −/− mice (n=8–10 per group) was assessed by quantitative real-time PCR. Levels of each target transcript are normalised against endogenous mRNA transcripts of the housekeeping gene Arbp and are expressed as arbitrary units (AU). Values are reported as means±SEM. Differences between fed and fasted conditions for each group and differences between groups (indicated by square brackets) were determined by two-way ANOVA followed by Fisher’s least significant differences post hoc analysis. *p<0.05; **p<0.01; ***p<0.001
We then assessed levels of Cd36 mRNA, which is translated into CD36, the putative long-chain fatty acid transporter of skeletal muscle [29] (Fig. 1). Fasting was associated with an increase in Cd36 expression in the wild-type (214%, p<0.001) and Tg-Prkag3 225Q (183%, p<0.001) mice (Fig. 2b). This fasting-induced increase in Cd36 expression was impaired in the absence of the AMPK γ3 subunit (70%, p<0.05). Overexpression of the mutant γ3225Q subunit was associated with an elevation in Cd36 expression in skeletal muscle during fasting.
Another rate-determining step in lipid metabolism is the transport of the fatty acid into the mitochondria as acyl-CoA. The transport of NEFAs from the sarcoplasm into the mitochondria is facilitated by the carnitine palmitoyl transfer system, which involves CPT-1 (Fig. 1). Cpt1 transcription was increased in response to fasting in wild-type (45%, p<0.01) and Tg-Prkag3 225Q (51%, p<0.01) mice, whereas the fasting-induced response was blunted in Prkag3 −/− mice (Fig. 2c).
In the mitochondrial matrix, acyl-CoA is oxidised in a stepwise manner by a series of enzymes, including HADHSC, leading to the formation of acetyl-CoA (Fig. 1). The transition from the fed to the fasted state was associated with a trend towards an increase in Hadhsc mRNA (41%, p=0.07) in wild-type mice, with essentially similar responses observed among the three groups (Fig. 2d). In the tricarboxylic acid (TCA) cycle, acetyl-CoA derived from oxidation of acyl-CoA or pyruvate is converted to citrate by CS, the first rate-controlling enzyme of the TCA cycle (Fig. 1). Fasting increased the expression of Cs mRNA in Tg-Prkag3 225Q mice (79%, p<0.01). In Prkag3 −/− mice, fasting also increased Cs expression (121%, p<0.001); however, the increase was lower compared with the pronounced effect (240%, p<0.001) noted in wild-type mice.
Electrons derived from the oxidation of substrate in the TCA cycle are transferred to the electron-transport chain to generate a proton gradient across the inner mitochondrial membrane for oxidative phosphorylation. The electron-transport chain is catalysed by a series of enzymes that require CYCS (Cycs) as a co-enzyme for shuttling electrons. Under fed conditions, levels of Cycs mRNA were greater in Tg-Prkag3 225Q mice than in wild-type (37%, p<0.05) or Prkag3 −/− mice (45%, p<0.05) (Fig. 2f). A similar increase in Cycs expression was observed under fasting conditions.
Under fasting conditions, expression of mitochondrial UCP3 is upregulated [4, 30]. One putative function of UCP3 is to export fatty acid anions from the mitochondrial matrix when acyl-CoA flux and lipid oxidation is increased [6, 31, 32] (Fig. 1). Fasting augmented Ucp3 expression (175%, p<0.001) in wild-type mice (Fig. 2g); this increase was more dramatic (328%, p<0.001) in Prkag3 −/− mice.
Genes implicated in glucose metabolism
Skeletal muscle glucose uptake is tightly regulated by GLUT4 (which is encoded by Slc2a4), a mammalian facilitative glucose transporter (Fig. 1). Slc2a4 mRNA expression was similar among the three groups. Upon entry into the cytoplasm, glucose is phosphorylated by HK2 to glucose 6-phosphate in the first rate-determining step of glycolysis (Fig. 1). Fasting induced a prominent decrease (77%, p<0.01) in Hk2 expression in Tg-Prkag3 225Q mice (Fig. 3b). Moreover, the level of Hk2 mRNA in Tg-Prkag3 225Q mice was markedly lower than that in wild-type mice (79%, p<0.05).
Relative expression of genes regulating glucose metabolism. The expression of Slc2a4 (a), Hk2 (b), Pfkm (c), Gys (d); Pdk4 (e) and Ldh2 (f), which encode proteins involved in regulating glucose metabolism, in white glycolytic gastrocnemius muscle of fed (white bars) and fasted (black bars) wild-type (WT), Tg-Prkag3 225Q and Prkag3 −/− mice (n=8–10 per group) was assessed by quantitative real-time PCR. Values of each target transcript are normalised against endogenous mRNA of the housekeeping gene Arbp and are expressed as arbitrary units (AU). Values are reported as means±SEM. Differences between fed and fasted conditions in each group and differences between groups (indicated by square brackets) were determined by two-way ANOVA followed by Fisher’s least significant differences post hoc analysis. *p<0.05; **p<0.01; ***p<0.001
Downstream of HK2, glucose can be further metabolised through glycolysis or glycogenesis (Fig. 1). We next determined the expression of PFKM, an enzyme that catalyses the second rate-controlling step of glycolysis. Fasting triggered a pronounced decrease (46%, p<0.001) in Pfkm expression in Tg-Prkag3 225Q mice, with no significant change in wild-type or Prkag3 −/− mice (Fig. 3c). Fasting increased (193%, p<0.001) the level of Gys mRNA in wild-type mice (Fig. 3d). This increase was less profound in Prkag3 −/− mice (98%, p<0.01), which expressed a lower level (29%, p<0.05) of this transcript than wild-type mice. Consistent with the reduced Hk2 expression, Gys expression was not elevated in Tg-Prkag3 225Q mice in response to fasting.
The link between glycolysis and the TCA cycle involves the decarboxylation of pyruvate to acetyl-CoA by pyruvate dehydrogenase. The activity of pyruvate dehydrogenase is downregulated when phosphorylated by PDK4 [33] (Fig. 1). Fasting drastically augments the expression of Pdk4 in skeletal muscle [4, 34, 35]. Moreover, the fasting-induced increase in Pdk4 is a proposed mechanism by which entry of pyruvate into the TCA cycle is inhibited to enhance the entry of acetyl-CoA derived from lipid oxidation [35]. Pdk4 mRNA was profoundly increased by fasting in wild-type mice (15-fold, p<0.001) (Fig. 3e), with a similar response noted in Tg-Prkag3 225Q (11-fold, p<0.001) and Prkag3 −/− (17-fold, p<0.001) mice. Thus, Pdk4 expression during fasting is unlikely to be regulated by AMPK heterotrimeric complexes containing the γ3 subunit.
We then evaluated whether fasting influences the expression of Ldh2 mRNA, another enzyme implicated in the pyruvate metabolism (Fig. 1). Fasting induced a two-fold (p<0.05) increase in Ldh2 expression in wild-type mice (Fig. 3f). However, this fasting response was blunted in the Prkag3 −/− mice, suggesting the obligatory role of AMPK γ3 subunit.
PPAR
Fasting induced a non-significant trend towards increased Ppara expression in wild-type mice. Ppard expression was decreased during fasting in Tg-Prkag3 225Q mice compared with that in wild-type (61%, p<0.01) and Prkag3 −/− (58%, p<0.05) mice (Fig. 4b). Conversely, the expression of Pparg was increased in the skeletal muscle of Tg-Prkag3 225Q mice under fed and fasting conditions (Fig. 4c).
Relative expression of the genes encoding the PPARs. Expression of Ppara (a), Ppard (b) and Pparg (c) in white glycolytic gastrocnemius muscle of fed (white bars) and fasted (black bars) wild-type (WT), Tg-Prkag3 225Q and Prkag3 −/− mice (n=8–10 per group) was assessed by quantitative real-time PCR. Values of each target transcript are normalised against endogenous mRNA of the housekeeping gene Arbp and are expressed as arbitrary units (AU). Values are reported as means±SEM. Differences between fed and fasted conditions for each group and differences between groups (indicated by squared brackets) were determined by two-way ANOVA followed by Fisher’s least significant differences post hoc analysis. *p<0.05; **p<0.01; ***p<0.001
Proteins involved in metabolism
Levels of CD36 were lower in Prkag3 −/− mice than in wild-type (38%, p<0.05) or Tg-Prkag3 225Q mice (40%, p<0.05) under fasting conditions. In contrast, levels of CD36 were increased in Tg-Prkag3 225Q mice under fed conditions (by 30% and 34% relative to those in wild-type and Prkag3 −/− mice, respectively, p<0.05). Levels of UCP3 were also increased in Tg-Prkag3 225Q mice (by 33% compared with those in Prkag3 −/− mice, p<0.05) (Fig. 5b). In contrast to the mRNA results, GLUT4 (Fig. 5c) and HK2 (Fig. 5d) levels were similar for the three groups and were unaltered by fasting. However, fasting increased GYS levels in wild-type (179%, p<0.05) and Prkag3 −/− mice (139%, p<0.05) (Fig. 5e). Consistent with the sustained elevation in glycogen content, fasting had no effect on GYS levels in Tg-Prkag3 225Q mice.
Production of proteins involved in metabolism. Level of CD36 (a), UCP3 (b), GLUT4 (c), HK2 (d) and GYS (e) in white glycolytic gastrocnemius muscle of fed (white bars) and fasted (black bars) wild-type (WT), Tg-Prkag3 225Q and Prkag3 −/− mice (n=5–6 per group) was assessed by western blot analysis. Data are expressed as percentage of WT under fed conditions. Values are reported as means±SEM. Differences between fed and fasted conditions for each group (*p<0.05) and differences between groups (†p<0.05) were determined by two-way ANOVA followed by Fisher’s least significant differences post hoc analysis
Glycogen and triglyceride content
Glycogen content in fed and fasted Tg-Prkag3 225Q mice was increased two-fold (p<0.05) compared with that in wild-type and Prkag3 −/− mice (Fig. 6a), consistent with our previous observations [26]. The increase in glycogen content can be partially attributed to the increase in the expression of genes encoding proteins that regulate lipid and oxidative metabolism (Fig. 2), leading to a glycogen-sparing effect that promotes carbohydrate storage. Fasting leads to significant decrease in skeletal muscle glycogen content in all three groups of mice. In contrast, fed and fasted intramuscular triglyceride content was similar among the three groups (Fig. 6b).
Muscle glycogen and triglyceride content. a Glycogen content in white glycolytic gastrocnemius muscle of wild-type (WT), Tg-Prkag3 225Q and Prkag3 −/− mice (n=5–12 per group) under fed (white bars) and fasted (black bars) conditions. b Triglyceride content in white gastrocnemius muscle of WT, Tg-Prkag3 225Q and Prkag3 −/− mice (n=8–13 per group) under fed (white bars) and fasted (black bars) conditions. Values are reported as means±SEM. Differences between fed and fasted conditions among each group (*p<0.05) and differences between groups († p<0.05) were determined by two-way ANOVA followed by Fisher’s least significant differences post hoc analysis
Discussion
The transcription of genes regulating metabolic pathways is tightly coordinated to allow efficient metabolic flux [36]. We provide evidence for a coordinated increase in the transcription of genes involved in lipid and glucose metabolism in skeletal muscle under fasting conditions. Moreover, the AMPK γ3 subunit plays an important role in this regulation. The transcription of specific genes involved in lipid (Lpl, Cd36 and Cpt1) and glucose (Gys and Ldh2) metabolism is impaired in AMPK γ3 subunit knockout mice. Conversely, the expression of genes involved in lipid metabolism (Cd36, Cycs and Ucp3) is enhanced in the skeletal muscle of Tg-Prkag3 225Q mice. Although post-translational modification (in particular, protein phosphorylation) is an efficient way of modulating metabolic homeostasis, recent studies have revealed the significant contribution of the transcriptional control of metabolism [37, 38]. Metabolic flux can be cohesively modulated through the coordinated transcription of multiple enzymes along diverse metabolic pathways [36, 39]. Consistent with this, a clear, coordinated increase in mRNA transcripts of genes regulating lipid/oxidative metabolism (Lpl, Cd36, Cpt1, Cs and Ucp3) and glucose metabolism (Gys, Pdk4 and Ldh2) was observed in wild-type mice during fasting.
A fasting-induced increase in mRNA levels of genes regulating the lipid metabolic pathway in skeletal muscle has been previously described [4–6], consistent with the observed increase in lipid metabolism in the tissue under fasting conditions. Given that SNF1 mediates transcriptional adaptations in yeast during glucose deprivation [1–3], we hypothesised that AMPK (a mammalian homologue of SNF1) may play a critical role in mediating a similar response in skeletal muscle during fasting. Genetic ablation of the AMPK γ3 subunit impaired the fasting-induced augmentation of Lpl, Cd36 and Cpt1 mRNA expression in skeletal muscle. Thus, the AMPK γ3 subunit plays a key role in mediating the transcriptional adaptation to fasting. Furthermore, ablation of the AMPK γ3 subunit also prevented the fasting-induced upregulation of Gys and Ldh2 transcription, consistent with previous findings that AMPK regulates the expression of glucose-responsive genes in hepatocytes [40] and islet beta cells [41]. Our results provide evidence for a role of AMPK in the regulation of the transcription of genes involved in lipid and glucose metabolism in white skeletal muscle.
We further validated our hypothesis that AMPK mediates transcriptional adaptations in response to fasting by skeletal muscle-specific overexpression of the mutant (R225Q) AMPKγ3 subunit [26]. We observed an increase in the transcript level of genes essential for lipid metabolism (Cd36) and oxidative metabolism (Cycs) in the skeletal muscle of Tg-Prkag3 225Q mice. Consistent with this, the protein content of CD36 and UCP3 was increased in Tg-Prkag3 225Q mice. Conversely, a coordinated decrease in the expression of genes regulating glucose metabolism (Hk2, Pfkm and Gys) was observed in the skeletal muscle of Tg-Prkag3 225Q mice under fasting conditions. In line with this, GYS levels in Tg-Prkag3 225Q mice were lower under fasting conditions. Our finding of a reciprocal relationship between the transcription of genes regulating glucose and lipid/oxidative metabolism is in contrast with previous findings of a concerted increase in the expression of genes along these metabolic pathways in AICAR-treated rodents [10, 42, 43]. The interpretation of previous studies that adopted chronic AICAR treatment as a means of chemically activating AMPK has been hindered by the non-specificity of the compound, which also activates other AMP-regulated enzymes [7, 44]. Moreover, the use of AICAR fails to provide insight into the role of distinct AMPK subunit isoforms as mediators of gene regulatory responses. Our genetic approach has provided direct evidence for the role of the AMPK γ3 subunit in orchestrating the coordinated transcriptional modulation of genes essential for lipid and glucose metabolic pathways in glycolytic skeletal muscle.
AMPK and PPARs are involved in mediating cellular adaptations to metabolic challenges. However, evidence is lacking in regard to the role of AMPK in regulating the transcription of the genes for the PPAR isoforms. Overexpression of the mutant R225Q γ3 subunit increases expression of Pparg in skeletal muscle under both fed and fasting conditions, but reduces Ppard expression under fasting conditions. Whether the inverse relationship between Pparg and Ppard under fasting conditions is direct or indirect remains unclear. However, PPARα and PPARδ subtypes have some redundant functions in skeletal muscle; thus, the δ isoform could compensate for a deficiency in the α isoform [45]. The coordinated expression and functional redundancy of the three PPAR subtypes in skeletal muscle requires further investigation.
Genetic ablation of the AMPK γ3 subunit in skeletal muscle rendered a metabolically inflexible phenotype, whereby the fasting-induced increase in transcription of genes involved in lipid and glucose metabolism was impaired. Nonetheless, intramuscular glycogen and triglyceride levels in Prkag3 −/− mice were comparable to those in wild-type mice. Thus, despite an impaired response to the fasting-induced transcription of genes essential for lipid metabolism, modulation of the glucose–fatty acid cycle in Prkag3 −/− mice during fasting could possibly be sustained, at least in part, by Pdk4 expression, in order to shunt the entry of glucose-derived acetyl-CoA into the TCA cycle. Interestingly, overexpression of the γ3225Q subunit rendered a metabolically flexible phenotype, partly due to an enhanced transcriptional coordination of genes controlling glucose and lipid/oxidative metabolism in skeletal muscle. The elevated skeletal muscle glycogen content in Tg-Prkag3 225Q mice may be the result of a reduction in glucose metabolism and an increase in lipid metabolism, leading to a glucose-sparing effect that promotes glycogen accumulation. Consistent with this notion, we have shown that Tg-Prkag3 225Q mice fed a high-fat diet displayed a lower intramuscular triglyceride content and a concomitant increase in fatty acid oxidation [26].
In conclusion, our results provide evidence that AMPK is involved in modulating a plethora of key genes essential for the regulation of lipid and glucose metabolism in glycolytic skeletal muscle. Furthermore, we demonstrate that AMPK complexes containing the γ3 subunit play a critical role in the transcriptional regulation of the ligand-activated nuclear receptors, namely PPARα, δ and γ.
Abbreviations
- AICAR:
-
5-aminoimidazole-4-carboximide-1-β-4-ribofuranoside
- ACC:
-
acetyl-CoA carboxylase
- AMPK:
-
AMP-activated protein kinase
- ARBP:
-
acidic ribosomal phosphoprotein PO
- CPT-1:
-
carnitine palmitoyl transferase-1
- CS:
-
citrate synthase
- CYCS:
-
cytochrome C
- GYS:
-
glycogen synthase
- HK2:
-
hexokinase II
- LDH2:
-
lactate dehydrogenase 2
- LPL:
-
lipoprotein lipase
- PDK4:
-
pyruvate dehydrogenase kinase 4
- PFKM:
-
muscle 6-phosphofructokinase
- PPAR:
-
peroxisome proliferator-activated receptor
- HADHSC:
-
short-chain 3-hydroxyacyl-CoA dehydrogenase
- SNF1:
-
sucrose non-fermenting protein kinase 1
- TCA:
-
tricarboxylic acid
- UCP3:
-
uncoupling protein 3
References
Hardie DG, Carling D, Carlson M (1998) The AMP-activated/Snf1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Ann Rev Biochem 67:821–855
Johnston M (1999) Feasting, fasting and fermenting: glucose sensing in yeast and other cells. Trends Genet 15:29–33
Hardie DG (2004) AMP-activated protein kinase: a key system mediating metabolic responses to exercise. Med Sci Sports Exerc 36:28–34
Pilegaard H, Saltin B, Neufer PD (2003) Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes 52:657–662
De Lange P, Ragni M, Silvestri E et al (2004) Combined cDNA array/RT-PCR analysis of gene expression profile in rat gastrocnemius muscle: relation to its adaptive function in energy metabolism during fasting. FASEB J 18:350–352
Hildebrandt AL, Neufer PD (2000) Exercise attenuates the fasting-induced transcriptional activation of metabolic genes in skeletal muscle. Am J Physiol 278:E1078–E1086
Hardie DG (2003) Mini review: the AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology 144:5179–5183
Kurth-Kraczek E, Hirshman M, Goodyear L, Winder W (1999) 5′ AMP-activated protein kinase activation causes GLUT4 translocation in skeletal muscle. Diabetes 48:1667–1671
Merrill GF, Kurth EJ, Hardie DG, Winder WW (1997) AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Physiol 273:E1107–E1112
Winder WW, Holmes BF, Rubink DS, Jensen EB, Chen M, Holloszy JO (2000) Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol 88:2219–2226
Song XM, Fiedler M, Galuska D et al (2002) 5-Aminoimidazole-4-carboxamide ribonucleoside treatment improves glucose homeostasis in insulin-resistant diabetic (ob/ob) mice. Diabetologia 45:56–65
Zong H, Ren JM, Young LH et al (2002) AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A 99:15983–15987
Putman CT, Kiricsi M, Pearcey J et al (2003) AMPK activation increases uncoupling protein-3 expression and mitochondrial enzyme activities in rat muscle without fibre type transitions. J Physiol (Lond) 551:169–178
Berger J, Moller DE (2002) The mechanisms of actions of PPARs. Annu Rev Med 53:409–435
Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ (2001) Nuclear receptors and lipid physiology: opening the X-files. Science 294:1866–1870
Leone TC, Weinheimer CJ, Kelly DP (1999) A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A 96:7473–7478
Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W (1999) Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 103:1489–1498
Rosen ED, Spiegelman BM (2001) PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem 276:37731–37734
Hevener AL, He W, Barak Y et al (2003) Muscle-specific Pparγ deletion causes insulin resistance. Nat Med 9:1491–1497
Wang YX, Lee CH, Tiep S et al (2003) Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 113:159–170
Mahlapuu M, Johansson C, Lindgren K et al (2004) Expression profiling of the gamma-subunit isoforms of AMP-activated protein kinase suggests a major role for gamma 3 in white skeletal muscle. Am J Physiol 286:E194–E200
Milan D, Jeon J-T, Looft C et al (2000) A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science 288:1248–1251
Estrade M, Vignon X, Rock E, Monin G (1993) Glycogen hyper-accumulation in white muscle fibres of RN-carrier pigs. A biochemical and ultrastructural study. Comp Biochem Physiol B 104:321–326
Estrade M, Ayoub S, Talmant A, Monin G (1994) Enzyme activities of glycogen metabolism and mitochondrial characteristics in muscles of RN-carrier pigs (Sus scrofa domesticus). Comp Biochem Physiol Biochem Mol Biol 108:295–301
Lebret B, Le Roy P, Monin G et al (1999) Influence of the three RN genotypes on chemical composition, enzyme activities, and myofiber characteristics of porcine skeletal muscle. J Anim Sci 77:1482–1489
Barnes BR, Marklund S, Steiler TL et al (2004) The 5′-AMP-activated protein kinase gamma3 isoform has a key role in carbohydrate and lipid metabolism in glycolytic skeletal muscle. J Biol Chem 279:38441–38447
Barnes BR, Glund S, Long YC, Hjalm G, Andersson L, Zierath JR (2005) 5′-AMP-activated protein kinase regulates skeletal muscle glycogen content and ergogenics. FASEB J 19:773–779
Wallberg-Henriksson H, Zetan N, Henriksson J (1987) Reversibility of decreased insulin-stimulated glucose transport capacity in diabetic muscle with in vitro incubation. Insulin is not required. J Biol Chem 262:7665–7671
Coburn CT, Knapp FF Jr, Febbraio M, Beets AL, Silverstein RL, Abumrad NA (2000) Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J Biol Chem 275:32523–32529
Gong D-W, He Y, Karas M, Reitman M (1997) Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta 3-adrenergic agonists, and leptin. J Biol Chem 272:24129–24132
Garvey WT (2003) The role of uncoupling protein 3 in human physiology. J Clin Invest 111:438–441
Himms-Hagen J, Harper M-E (2001) Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: a hypothesis. Exp Biol Med 226:78–84
Sugden MC, Holness MJ (2003) Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol 284:E855–E862
Wu P, Inskeep K, Bowker-Kinley M, Popov K, Harris R (1999) Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes 48:1593–1599
Sugden MC, Kraus A, Harris RA, Holness MJ (2000) Fibre-type specific modification of the activity and regulation of skeletal muscle pyruvate dehydrogenase kinase (PDK) by prolonged starvation and refeeding is associated with targeted regulation of PDK isoenzyme 4 expression. Biochem J 346:651–657
Ihmels J, Levy R, Barkai N (2004) Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol 22:86–92
DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686
Miki R, Kadota K, Bono H et al (2001) Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc Natl Acad Sci U S A 98:2199–2204
Kacser H, Acerenza L (1993) A universal method for achieving increases in metabolite production. Eur J Biochem 216:361–367
Woods A, Azzout-Marniche D, Foretz M et al (2000) Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Mol Cell Biol 20:6704–6711
Da Silva Xavier G, Leclerc I, Salt IP et al (2000) Role of AMP-activated protein kinase in the regulation by glucose of islet beta cell gene expression. Proc Natl Acad Sci U S A 97:4023–4028
Holmes BF, Kurth-Kraczek EJ, Winder WW (1999) Chronic activation of 5′-AMP-activated protein kinase increases GLUT-4, hexokinase, and glycogen in muscle. J Appl Physiol 87:1990–1995
Stoppani J, Hildebrandt AL, Sakamoto K, Cameron-Smith D, Goodyear LJ, Neufer PD (2002) AMP-activated protein kinase activates transcription of the UCP3 and HKII genes in rat skeletal muscle. Am J Physiol 283:E1239–E1248
Kemp BE, Mitchelhill KI, Stapleton D, Michell BJ, Chen Z-P, Witters LA (1999) Dealing with energy demand: the AMP-activated protein kinase. Trends Biochem Sci 24:22–25
Muoio DM, MacLean PS, Lang DB et al (2002) Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. J Biol Chem 277:26089–26097
Acknowledgements
This work was supported by grants from the Swedish Medical Research Council, the Swedish Diabetes Association, the Novo-Nordisk Research Foundation, the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning and the Swedish Foundation for Strategic Research.
Duality of interest. M. Mahlapuu and S. Martinsson are employed by Arexis (Göteborg, Sweden).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Long, Y.C., Barnes, B.R., Mahlapuu, M. et al. Role of AMP-activated protein kinase in the coordinated expression of genes controlling glucose and lipid metabolism in mouse white skeletal muscle. Diabetologia 48, 2354–2364 (2005). https://doi.org/10.1007/s00125-005-1962-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-005-1962-5