Abstract
Key message
Quantitative trait locus analysis identified independent novel loci in cucumbers responsible for resistance to races 0 and 1 of the anthracnose fungal pathogen Colletotrichum orbiculare.
Abstract
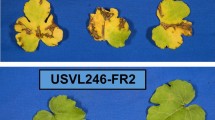
Cucumbers have been reported to be vulnerable to Colletotrichum orbiculare, causing anthracnose disease with significant yield loss under favorable conditions. The deployment of a single recessive Cssgr gene in cucumber breeding for anthracnose resistance was effective until a recent report on high-virulent strains infecting cucumbers in Japan conquering the resistance. QTL mapping was conducted to identify the resistance loci in the cucumber accession Ban Kyuri (G100) against C. orbiculare strains 104-T and CcM-1 of pathogenic races 0 and 1, respectively. A single dominant locus An5 was detected in the disease resistance hotspot on chromosome 5 for resistance to 104-T. Resistance to CcM-1 was governed by three loci with additive effects located on chromosomes 2 (An2) and 1 (An1.1 and An1.2). Molecular markers were developed based on variant calling between the corresponding QTL regions in the de novo assembly of the G100 genome and the publicly available cucumber genomes. Multiple backcrossed populations were deployed to fine-map An5 locus and narrow the region to approximately 222 kbp. Accumulation of An2 and An1.1 alleles displayed an adequate resistance to CcM-1 strain. This study provides functional molecular markers for pyramiding resistance loci that confer sufficient resistance against anthracnose in cucumbers.





Similar content being viewed by others
Data availability
The supporting data for this study are available in the Supplementary Material. The nucleotide sequence data reported herein are available from the DNA Data Bank of Japan (DDBJ) Sequence Read Archive. The other datasets generated in this study are available from the corresponding author upon request.
References
Abul-Hayja Z, Willams P, Peterson C (1978) Inheritance of resistance to anthracnose and target leaf spot in cucumbers. Plant Dis Rep 62:43–45
Arends D, Prins P, Jansen RC, Broman KW (2010) R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26:2990–2992. https://doi.org/10.1093/bioinformatics/btq565
Barnes WC, Epps WM (1952) A unreported type of resistance to cucumber downy mildew. Plant Dis Rep 38:620
Barnes W, Epps W (1954) Two types of anthracnose resistance in cucumbers. Plant Dis Rep 36:479–480
Berg J, a, Appiano M, Santillán Martínez M, et al (2015) A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol 15:243. https://doi.org/10.1186/s12870-015-0635-x
Berg JA, Hermans FWK, Beenders F et al (2020) Analysis of QTL DM4.1 for downy mildew resistance in cucumber reveals multiple subQTL: A novel RLK as candidate gene for the most important subQTL. Front Plant Sci 11:1–18. https://doi.org/10.3389/fpls.2020.569876
Bergelson J, Kreitman M, Stahl EA et al (2001) Evolutionary dynamics of plant R-genes. Science 292:2281–2285. https://doi.org/10.1126/science.1061337
Busch L, Walker J (1958) Studies of cucumber anthracnose. Phytopathology 48:302–304
Chen Y, Nie F, Xie S-Q et al (2021) Efficient assembly of nanopore reads via highly accurate and intact error correction. Nat Commun 12:60. https://doi.org/10.1038/s41467-020-20236-7
De Beukelaer H, Davenport GF, Fack V (2018) Core Hunter 3: flexible core subset selection. BMC Bioinformatics 19:203. https://doi.org/10.1186/s12859-018-2209-z
Fanourakis NE, Simon PW (1987) Analysis of genetic linkage in the cucumber. J Hered 78:1422–1427
Goode MJ (1956) Physiologic specialization in Colletotrichum lagenarium. Plant Dis Rep 40:741
Inoue Y, Phuong Vy TT, Singkaravanit-Ogawa S et al (2023) Selective deployment of virulence effectors correlates with host specificity in a fungal plant pathogen. New Phytol 238:1578–1592. https://doi.org/10.1111/nph.18790
Jenkins S, Winstead N, McCombs C (1964) Pathogenic comparison of three new and four previously described races of Glomerella angulata var. orbiculare. Plant Dis Rep 48:619–623
Kang H, Weng Y, Yang Y et al (2011) Fine genetic mapping localizes cucumber scab resistance gene Ccu into an R gene cluster. Theor Appl Genet 122:795–803. https://doi.org/10.1007/S00122-010-1487-2/FIGURES/3
Kelly JD, Vallejo V (2006) QTL analysis of multigenic disease resistance in plant breeding. In: Tuzun S, Bent E (eds) Multigenic and Induced Systemic Resistance in Plants. Springer, US, pp 21–48
Kourelis J, Van Der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30:285–299. https://doi.org/10.1105/tpc.17.00579
Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439
Kubo Y (2018) Infection structure development and pathogenesis of Colletotrichum orbiculare. Jpn J Phytopathol 84:131–134. https://doi.org/10.3186/jjphytopath.84.131
Kubo Y, Harata K, Kodama S, Fukada F (2016) Development of the infection strategy of the hemibiotrophic plant pathogen, Colletotrichum orbiculare, and plant immunity. Physiol Mol Plant Pathol 95:32–36. https://doi.org/10.1016/j.pmpp.2016.02.008
Kubo Y, Takano Y (2013) Dynamics of infection-related morphogenesis and pathogenesis in Colletotrichum orbiculare. J Gener Plant Pathol
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Method 9(4–9):357–359. https://doi.org/10.1038/nmeth.1923
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Li S, Wang H, Huo Z, Guan W (2008) Development of SCAR marker linked to cucumber anthracnose resistance-related gene from an AFLP marker. Acta Hort 35:123–126 ((in Chinese))
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li Q, Li H, Huang W et al (2019) A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). GigaScience 8:1–20. https://doi.org/10.1093/GIGASCIENCE/GIZ072
Linde D, Bridges W, Rhodes B (1990) Inheritance of resistance in cucumber to race 2 of Colletotrichum lagenarium. Theor Appl Genet 79:12–16
Ma M, Yang L, Hu Z, Mo C et al (2024) Multiplex gene editing reveals cucumber MILDEW RESISTANCE LOCUS O family roles in powdery mildew resistance. Plant Physiol 00:1–20. https://doi.org/10.1093/plphys/kiae047
Matsuo H, Ishiga Y, Kubo Y, Yoshioka Y (2022a) Colletotrichum orbiculare strains distributed in Japan: race identification and evaluation of virulence to cucurbits. Breed Sci 72:22011. https://doi.org/10.1270/jsbbs.22011
Matsuo H, Sugiyama M, Yoshioka Y (2022b) Identification of a new source of resistance to anthracnose in cucumber in Japan. Hortic J 91:49–57. https://doi.org/10.2503/HORTJ.UTD-322
Morata J, Puigdomènech P (2017) Variability among Cucurbitaceae species (melon, cucumber and watermelon) in a genomic region containing a cluster of NBS-LRR genes. BMC Genomics 18:1–7. https://doi.org/10.1186/s12864-017-3529-5
Naito K (2023) How to sewuence and assemble plant genomes. In: Arakawa K (ed) Nanopore Sequencing: Methods and Protocols. Springer Nature, US, pp 57–78
Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18:613–615. https://doi.org/10.1016/S0168-9525(02)02820-2
Nie J, Wang Y, He H et al (2015) Loss-of-function mutations in CsMLO1 confer durable powdery mildew resistance in cucumber (Cucumis sativus L.). Front Plant Sci 6:1–14. https://doi.org/10.3389/fpls.2015.01155
Ohta S, Yano K, Kurita Y et al (2013) A sample preparation method for direct and non-direct PCR in woody plants. J Jpn Soc Hortic Sci 82:14–21. https://doi.org/10.2503/jjshs1.82.14
Ouellette LA, Reid RW, Blanchard SG, Brouwer CR (2018) LinkageMapView-rendering high-resolution linkage and QTL maps. Bioinform (oxf, Eng) 34:306–307. https://doi.org/10.1093/BIOINFORMATICS/BTX576
Pan J, Tan J, Wang Y et al (2018) STAYGREEN (CsSGR) is a candidate for the anthracnose (Colletotrichum orbiculare) resistance locus cla in Gy14 cucumber. Theor Appl Genet 131:1577–1587. https://doi.org/10.1007/s00122-018-3099-1
Pan Y, Wang Y, McGregor C et al (2020) Genetic architecture of fruit size and shape variation in cucurbits: a comparative perspective. Theor Appl Genet 133:1–21. https://doi.org/10.1007/s00122-019-03481-3
Pilet-Nayel M-L, Moury B, Caffier V et al (2017) Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front Plant Sci 8:1838. https://doi.org/10.3389/fpls.2017.01838
Sacristán S, Goss EM, den Akker SE (2021) How Do Pathogens Evolve Novel Virulence Activities? Am Phytopathol Soc 34:576–586. https://doi.org/10.1094/MPMI-09-20-0258-IA
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. https://doi.org/10.1093/BIOINFORMATICS/BTR026
Seiko T, Muto C, Shimomura K et al (2024) Chromosome-level assembly of Cucumis sativus cv. ‘Tokiwa’ as a reference genome of Japanese cucumber. bioRxiv. https://doi.org/10.1101/2024.04.15.589484
Shimomura K, Sugiyama M, Kawazu Y, Yoshioka Y (2021a) Quantitative trait locus analysis of cucumber fruit texture using double-digest restriction-site-associated DNA sequencing. Euphytica 217:1–14. https://doi.org/10.1007/s10681-021-02830-y
Shimomura K, Sugiyama M, Kawazu Y, Yoshioka Y (2021b) Identification of quantitative trait loci for powdery mildew resistance in highly resistant cucumber (Cucumis sativus L.) using ddRAD-seq analysis. Breed Sci 71:326–333. https://doi.org/10.1270/jsbbs.20141
Shirasawa K, Hirakawa H, Isobe S (2016) Analytical workflow of double-digest restriction site-associated DNA sequencing based on empirical and in silico optimization in tomato. DNA Res: an Int J Rapid Publ Rep Genes Genom 23:145–153. https://doi.org/10.1093/DNARES/DSW004
Sitterly W (1973) Cucurbits. In: Nelson RR (ed) Breeding plants for disease resistance, concepts and applications. Penn State University Press, University Park, pp 287–306
Takano Y, Inoue Y, Zhang R et al (2023) Nonhost resistance and effectors in interactions between Colletotrichum species and plants. Physiol Mol Plant Pathol 125:101982. https://doi.org/10.1016/j.pmpp.2023.101982
Tan J, Wang Y, Dymerski R et al (2022) Sigma factor binding protein 1 (CsSIB1) is a putative candidate of the major-effect QTL dm5.3 for downy mildew resistance in cucumber (Cucumis sativus). Theor Appl Genet 135:4197–4215. https://doi.org/10.1007/s00122-022-04212-x
Taylor J, Butler D (2017) R package ASMap: efficient genetic linkage map construction and diagnosis. J Stat Software 79(6):1–29. https://doi.org/10.18637/jss.v079.i06
Tek MI, Calis O, Fidan H et al (2022) CRISPR/Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front Plant Sci 13:1081506. https://doi.org/10.3389/fpls.2022.1081506
Thompson D, Jenkins S (1985) Influence of cultivar resistance, initial disease, environment, and fungicide concentration and timing on anthracnose development and yield loss in pickling cucumbers. Phytopathology 75:1422–1427
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115–e115. https://doi.org/10.1093/nar/gks596
Vaser R, Sović I, Nagarajan N, Šikić M (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. https://doi.org/10.1101/gr.214270.116
Wan H, Yuan W, Bo K et al (2013) Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. https://doi.org/10.1186/1471-2164-14-109
Wang HJ, Li S, Liu X et al (2007) AFLP markers of cucumber anthracnose resistance-related gene. Acta Hort 34:213–216 ((in Chinese))
Wang Y, Tan J, Wu Z et al (2019) STAYGREEN, STAY HEALTHY: a loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol 221:415–430. https://doi.org/10.1111/nph.15353
Wang Y, Bo K, Gu X et al (2020) Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic Res 7:1–20. https://doi.org/10.1038/s41438-019-0226-3
Wang Y, VandenLangenberg K, Wehner TC, Weng Y (2014) QTLs for downy mildew resistance and their association with LRR-RLK / RLP resistance gene homologs in cucumber. In: Cucubitaceae 2014. Michigan, USA
Wasilwa LA, Correl JC, Morelock TE, McNew RE (1993) Reexamination of races of the cucurbit anthracnose pathogen Colletorichum orbiculare. Phytopathology 83:1190–1198
Wen C, Mao A, Dong C et al (2015) Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber, Cucumis sativus L. Theor Appl Genet 128:2495–2506. https://doi.org/10.1007/S00122-015-2604-z
Wyszogrodzka A, Williams P, Peterson C (1987) Multiple-pathogen inoculation of cucumber (Cucumis sativus) seedlings. Plant Dis 71:275–280
Xu X, Wang W, Du Y et al (2023) A single-nucleotide substitution in the leucine-rich-repeat-only gene CsLRR1 confers powdery mildew resistance in cucumber. Plant Commun. https://doi.org/10.1016/j.xplc.2023.100774
Yang L, Li D, Li Y et al (2013) A 1,681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol 13:1–14. https://doi.org/10.1186/1471-2229-13-53
Yu G, Chen Q, Wang X et al (2019) Mildew resistance locus o genes CsMLO1 and CsMLO2 are negative modulators of the Cucumis sativus defense response to Corynespora cassiicola. Int J Mol Sci 20:1–24. https://doi.org/10.3390/ijms20194793
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468. https://doi.org/10.1093/GENETICS/136.4.1457
Zhang S, ping, Miao H, Yang Y hong, et al (2014) A major quantitative trait locus conferring resistance to fusarium wilt was detected in cucumber by using recombinant inbred lines. Mol Breed 34:1805–1815. https://doi.org/10.1007/S11032-014-0140-1/TABLES/3
Zhang K, Wang X, Zhu W et al (2018) Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor Appl Genet 131:2229–2243. https://doi.org/10.1007/S00122-018-3150-2
Zhang C, Badri Anarjan M, Win KT et al (2021) QTL-seq analysis of powdery mildew resistance in a Korean cucumber inbred line. Theor Appl Genet 134:435–451. https://doi.org/10.1007/s00122-020-03705-x
Zhang W, Yuan Q, Wu Y et al (2022) Genome-wide identification and characterization of the CC-NBS-LRR gene family in cucumber (Cucumis sativus L.). Int J Mol Sci 23:5048–5048. https://doi.org/10.3390/IJMS23095048
Acknowledgements
We thank Z. Shuang, N. Nishi, and K. Takano of the University of Tsukuba for their technical assistance. Fungal strains and plant genetic resources were obtained from Genebank of the National Agriculture and Food Research Organization (NARO), Japan. This work was partly supported by the Ministry of Agriculture, Forestry and Fisheries (MAFF) of the Government of Japan commissioned project “A Collaborative Research Project on Characterization and Evaluation of Plant Genetic Resources for Food and Agriculture (PGRAsia)” (grant no. JPJ007117), the Japan Science and Technology Agency (Support for Pioneering Research Initiated by the Next Generation (SPRING); Grant number: JPMJSP2124), Grant-in-Aid for JSPS Fellows, JSPS KAKENHI Grant Number 23KJ1148.
Funding
This work was partly supported by the Ministry of Agriculture, Forestry, and Fisheries (MAFF) of the Government of Japan commissioned project ‘A Collaborative Research Project on Characterization and Evaluation of Plant Genetic Resources for Food and Agriculture (PGRAsia)’ (grant no. JPJ007117), the Japan Science and Technology Agency (Support for Pioneering Research Initiated by the Next Generation (SPRING); Grant number: JPMJSP2124), Grant-in-Aid for JSPS Fellows, JSPS KAKENHI Grant Number 23KJ1148.
Author information
Authors and Affiliations
Contributions
YY designed and supervised the study. FF and HM developed the populations and evaluated their phenotypes. SI and KS performed ddRAD sequencing. HM built a linkage map and performed QTL analysis. FF developed DNA markers and performed fine mapping and marker analysis. KN performed the sequencing and assembly of the G100 genome. FF and HM prepared the manuscript. All the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest relevant to the content of this article.
Additional information
Communicated by Yiqun Weng.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fitriyah, F., Matsuo, H., Isobe, S. et al. Different control of resistance to two Colletotrichum orbiculare pathogenic races 0 and 1 in cucumber (Cucumis sativus L.). Theor Appl Genet 137, 127 (2024). https://doi.org/10.1007/s00122-024-04633-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-024-04633-w