Abstract
Key Message
By integrating QTL mapping, transcriptomics and metabolomics, 138 hub genes were identified in rapeseed root response to aluminum stress and mainly involved in metabolism of lipids, carbohydrates and secondary metabolites.
Abstract
Aluminum (Al) toxicity has become one of the important abiotic stress factors in areas with acid soil, which hinders the absorption of water and nutrients by roots, and consequently retards the growth of crops. A deeper understanding of the stress-response mechanism of Brassica napus may allow us to identify the tolerance gene(s) and use this information in breeding-resistant crop varieties. In this study, a population of 138 recombinant inbred lines (RILs) was subjected to aluminum stress, and QTL (quantitative trait locus) mapping was used to preliminarily locate quantitative trait loci related to aluminum stress. Root tissues from seedlings of an aluminum-resistant (R) line and an aluminum-sensitive (S) line from the RIL population were harvested for transcriptome sequencing and metabolome determination. By combining the data on quantitative trait genes (QTGs), differentially expressed genes (DEGs), and differentially accumulated metabolites (DAMs), key candidate genes related to aluminum tolerance in rapeseed were determined. The results showed that there were 3186 QTGs in the RIL population, 14,232 DEGs and 457 DAMs in the comparison between R and S lines. Lastly, 138 hub genes were selected to have a strong positive or negative correlation with 30 important metabolites (|R|≥ 0.95). These genes were mainly involved in the metabolism of lipids, carbohydrates and secondary metabolites in response to Al toxicity stress. In summary, this study provides an effective method for screening key genes by combining QTLs, transcriptome sequencing and metabolomic analysis, but also lists key genes for exploring the molecular mechanism of Al tolerance in rapeseed seedling roots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum toxicity is a serious problem of crop production in acidic soils, which constitute about 50% of the world’s potentially arable lands (Kochian et al. 2005). In recent decades, the acidification of soil has become more problematic because of the excessive application of ammonium fertilizer and the increase in acid rain. Studies have proven that excess Al3+ content in acid soil can inhibit cellular activity in root meristem, prevent mitosis and retard root growth (Doncheva et al. 2005; Marciano et al. 2010). A healthy root system is crucial for providing anchorage, the intake of water and nutrients, sensing environmental signals and interacting with the soil microbiome (Tracy et al. 2020). Root system growth is regulated by both genetic and environmental factors (Slovak et al. 2016; Waidmann et al. 2020), and crop germplasm shows extensive genetic variation to adverse stress including aluminum tolerance. Root adaptations can enhance the plant`s resilience to Al3+ stress; therefore, root systems are favorable targets for potential improvement in plant growth and crop productivity (Godfray et al. 2010). The mechanisms of plant resistance to aluminum stress are complex and diverse, including secretion of organic acids, increase of rhizosphere pH, secretion of mucus, fixation of Al3+ in cell walls, chelation of Al3+ by organic acids in solution and isolation in vacuolar areas (Kochian et al. 2015). In summary, plants’ Al resistance mechanisms can be categorized according to whether they occur within or outside of the plant cell as external exclusion or internal detoxification (Wang et al. 2021a, b). With regard to the external exclusion mode of Al3+ toxicity, the best-studied process is that of secretion of organic acid ions such as citrate, oxalate or malate, from the root tip, which can then directly chelate external Al3+ and prevent toxicity (Lou et al. 2020; Ma et al. 2001; Yang et al. 2019). Most studies have focused on analyzing conventional physiological characteristics, but in-depth molecular biological analyses are lacking. Understanding the molecular mechanism underlying the regulation of root development of rapeseed to Al3+ stress will help us by facilitating the application of genetic engineering strategies and chemical regulation to improve aluminum tolerance in crop plants.
Quantitative trait locus (QTL) mapping is an effective method to identify aluminum tolerance genes (Bian et al. 2013). Many quantitative trait loci (QTLs) associated with Al3+ toxicity have been detected in a variety of crops, including wheat (Farokhzadeh et al. 2020), soybean (Li et al. 2021) and rice (Jahan et al. 2021). Consequently, QTL mapping generates a number of candidate genes due to the small population used and the low density of the genetic map (Mackay et al. 2009). High-throughput RNA sequencing (RNA-seq) technology has become a powerful tool. It offers the benefits of high sensitivity and cost-effectiveness, and it has been used to study Al3+ stress-induced changes in the genomes of rice (Awasthi et al. 2021), peanuts (Xiao et al. 2021) and tea (Huang et al. 2021). Similarly, transcriptome analysis can identify thousands of differentially expressed genes (DEGs) associated with Al3+ stress; however, it is difficult to identify all the key genes related to aluminum stress. Therefore, the integration of DEG identification with QTL linkage mapping has been considered to be the most effective way to delineate candidate genes related to complex traits. This approach has been applied to rice (Lei et al. 2020), maize (Han et al. 2022), alfalfa (Jiang et al. 2022) and grapes (Ma et al. 2022). In addition, it was also used in the determination of resistance to Al3+ stress during rapeseed germination (Wang et al. 2021a, b), resistance to horseshoe root disease (Kopec et al. 2021) and resistance to low temperature stress (Qin et al. 2022). Advances in transcriptome sequencing technology (RNA-seq) and metabolomics have enhanced their application in studying the response mechanism of plants to biotic and abiotic stress, and organically linked the metabolic pathways involved in differential gene expression with differentially accumulated metabolites (DAMs) (Chen et al. 2019; Meng et al. 2022; Zhao et al. 2019). Therefore, the application of multi-omics could be considered a fruitful approach for the initial discovery of key genes related to aluminum toxicity and the associated mechanisms at the molecular level.
Brassica napus (genome AACC, 2n = 38) is one of the most productive oilseed crops and an important source of protein-rich livestock feed around the world. It is mainly planted in the Yangtze River basin in China where the soil is more acidic, but aluminum toxicity has become one of the main limiting factor affecting rapeseed production in this region (Gao et al. 2021). In this study, root phenotypes, including relative root length (RRL), relative root diameter (RRD), relative root surface area (RSA), relative root volume (RRV), relative root tips (RRT) and relative dry weight (RDW), were investigated in rapeseed seedlings from the recombinant inbred line (RIL) population (ZS11 × 10D130) after exposure to Al3+. Next, QTLs were preliminarily located using the constructed genetic linkage map (Ye et al. 2019). Based on the gene annotation information of the reference genome of Brassica napus, quantitative trait genes (QTGs) were screened within mapping confidence intervals. Transcriptome sequencing (RNA-seq) was performed on an Al3+-resistant (R) line and an Al3+-sensitive (S) line from the RIL population to identify differentially expressed genes (DEGs), and genes shared by QTGs and DEGs were screened. According to the correlation between these genes and differentially accumulated metabolites (DAMs), the number of candidate genes was further narrowed. We employed a combinatorial approach of QTL mapping in a RIL population, RNA-sequence and metabolomics analysis of R and S lines to identify the key candidate genes governing root traits in rapeseed seedlings exposed to Al3+ stress.
Materials and methods
Experimental material
The RIL containing 138 lines and its parents were used for QTL mapping, and the Al3+-resistant line 27,034 (R) and the Al3+-sensitive line 27,143 (S) derived from the RIL population were used for RNA-seq, metabolomic analysis and physiological indicators determination. The RIL population came from a cross between 10D130 and Zhongshuang11 (ZS11) by the single-grain transmission method for seven consecutive generations. The 10D130 strain is a high-generation, Al3+-sensitive inbred line selected from interspecific hybrids of Brassica juncea and Brassica oleracea by the Chongqing Engineering Research Center, while ZS11 is an Al3+-resistant, conventional, high-quality rapeseed variety selected by the Chinese Academy of Agricultural Sciences. All seeds were provided by the Chongqing Engineering Research Center.
Experimental treatments and phenotypic evaluation
Full and uniform seeds selected from 138 lines of an RIL population were surface-sterilized, rinsed with deionized water and germinated in vermiculite medium. When the seedlings showed uniform growth with 2–3 true leaves, they were transferred to a 45-hole foam seedling tray (60 × 34 × 3 cm) and placed in a plastic tray (67 × 41 × 15 cm) for hydroponic culture in 1/2 Hoagland’s nutrient solution, in a climate-controlled chamber at 25 °C for 16 h during the day, and 20 °C for 8 h at night until four true leaves developed. The seedlings were subjected to Al stress in 1/2 Hoagland’s nutrient solution with 2.40 mM AlCl3·6H2O (pH 4.0) (Wu et al. 2022); 1/2 Hoagland’s nutrient solution without Al3+ was used as control. The treatment solutions were replaced every four days. After 15 days of treatment, the root systems were analyzed using the LD-WinRhizo scope (Regent Instruments, QC, Canada) to measure RRL, RRD, RRV, RSA, RRTs, and RDW. Each test was conducted with three replicates. In addition, mixed root samples from each biological replicate of the R line were collected into thirty 1.5 ml microfuge tubes, each containing 100 mg of root sample, quickly frozen in liquid nitrogen and stored at − 80 °C for transcriptome sequencing, identification of metabolites, determination of physiological indices and qRT-PCR. The same method was used to collect root samples for the S line. The control and treated samples of the R and S lines were labeled RCK and SCK, and RT and ST, respectively.
Determination of physiological parameters in roots
The activity of superoxide dismutase (SOD) was determined by xanthine oxidase cytochrome C. The activity of peroxidase (POD) was measured by the guaiacol method, and catalase (CAT) activity was determined by an ultraviolet absorption method. Soluble proteins (SP) were analyzed according to Sedmak and Grossberg (1977). The content of malondialdehyde (MDA) was determined by the thiobarbituric acid (TBA) reaction method. The content of Al3+ ions in the roots was measured by the method of Wright et al. (1987). The contents of SOD, POD, CAT, SP, MDA and Al3+ were assayed using kits according to manufacturer’s instructions (Suzhou Grace Biotechnology Co., Ltd, Suzhou, China). All measurements were performed in triplicate, and means ± SD were calculated.
QTL localization via interval gene (QTGs) screening
Based on a genetic linkage map constructed earlier in our laboratory (Ye et al. 2019), Map QTL6.0 software was used to detect QTL sites related to RRL, RRD, RRV, RSA, RRT and RDW. The logarithm of the odds (LOD) threshold for a significant QTL was calculated with 1000 permutations at a significance level of p = 0.05. The confidence interval for each QTL was defined by LOD ≥ 2 from the peak position. QTLs were assigned names by adding the prefix of the trait abbreviations, and their locations, including the chromosome number, as determined according to McCouch et al. (1997). If more than one QTL was detected on the same chromosome for a trait, the QTLs were consecutively numbered. MapChart 2.5 software was used to depict the QTL positions.
RNA sequencing (RNA-seq) and DEGs determination
The root samples of R and S lines from controls and Al3+ stress treatments were sent to Biomarker Technologies Co., Ltd (Beijing, China) for RNA extraction, library construction, and transcriptome sequencing on the Illumina sequencing platform. After removing the 3’-adapter, low-quality sequences and clean reads were aligned to the Brassica napus reference genome (https://www.genoscope.cns.fr/brassicanapus/cgi-bin/gbrowse/colza/) using HISAT2 software (http://ccb.jhu.edu/software/hisat2/index.shtml). The read count value was determined by HTSeq (https://htseq.readthedocs.io/en/release_0.11.1/). Fragments per kilobase million (FPKM) values were calculated to estimate gene expression levels. DESeq2 was used for differential expression analysis between the two groups based on p ≤ 0.01 and |log2 fold change|≥ 2. Enrichment analysis used the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway tool. TBtools software (Chen et al. 2020) was used to draw the Venn diagram and candidate gene expression heatmap.
Metabolite profiling
Twelve root samples from the R and S lines (T, CK) were used for untargeted metabolites based on ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS) (Biomarker Technologies Co., Ltd.). UPLC-Q-TOF/MS analyses were performed with an Acquisition I-Class plus tandem Xevo G2-XS QToF (Waters, Milford, USA). An Acquity UPLC HSS T3 column (Waters, Milford, USA) was used in this study. The samples were analyzed in both the positive and negative ion modes. Raw mass-spec data were collected using MassLynx software (version 4.2, Waters, Milford, USA), processed and analyzed by the Progenesis QI software (Waters, Milford, USA). The MetLin database (Waters, Milford, USA) and Biomark’s self-built database were used for peak annotation and identification. Simca-P software (version 14.1, Umetrics, Umeå, Sweden) was used for principal component analysis (PCA).
Candidate gene predication
QTL intervals were aligned to the Brassica napus reference genome (http://www.genoscope.cns.fr/brassicanapus/cgi-bin/gbrowse/colza/). Based on the physical positions of the flanking markers, the genes located within the QTL confidence interval were extracted (QTGs). The R language Hmisc package function (Harrell 2022) was used to determine the correlations of DEGs screened after combining with QTL and transcriptome data. The prediction of candidate genes was based on two conditions: whether the expression trends of the QTL confidence interval genes (QTGs) were different between RT vs. ST and RCK vs. SCK, and whether the expression trends of RT vs. RCK and ST vs. SCK were significantly different. Genes and metabolites with Pearson's correlation coefficients, |R|≥ 0.95 and p ≤ 0.01, were used to establish the related network, which was visualized using Cytoscape 3.8.0 software (Shannon et al. 2003).
Validation of candidate genes by qRT–PCR
Quantitative RT-PCR was performed on a BioRad CFX96 real-time system using a kit from Labgic Biotechnology Co., Ltd (Beijing, China). The primer pairs were designed by qPrimerDB v1.2 (https://biodb.swu.edu.cn/qprimerdb/) and synthesized by Sangon Biotech (Shanghai, China) (Table S1). The reaction conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 56 °C for 30 s and 72 °C for 30 s. BraActin7 was used as an internal control. The 2−ΔΔCt method was used to calculate the normalized expression of target genes. Three biological replicates per sample were analyzed for expression levels.
Data processing and analysis
SPSS 22.0 (SPSS Inc., Chicago, USA) was used for statistical testing of phenotypic data from the RIL population. Statistical analysis used DPS 6.0 (Analytical Software, Hangzhou, Zhejiang, China).
Results
Root growth of parents and RIL populations under Al3+ stress
As shown in Table 1, except for RRD and RDW, the root-related traits of the RIL population were inhibited by stress from 2.40 µM AlCl3·6H2O. In addition, the RRD in the S line was 1.017, which was less than the 1.284 of the R line. The other root-related traits in the S line ranged from 0.343 to 0.487, while those in the R line ranged from 0.651 to 0.906. This indicated that the tolerance of S and R lines to aluminum stress was significantly different from each other; the root growth in the S line was more severely inhibited than that of the R line after exposure to Al stress.
Frequency analysis showed that all six traits had a continuous distribution with extensive genetic variation (Fig. 1), which was in accordance with quantitative traits and was suitable for QTL mapping.
QTL linkage mapping of root phenotypes
Genotype analysis of the RIL population was conducted based on 5058 SNP markers (Table S2). In total, 17 QTLs significantly associated with aluminum stress were identified, including 4 related to RRL, 8 to RRD, 2 to RRT, 1 to RSA, 1 to RRV and 1 to RDW (Fig. 2). These QTLs explained 7.00–14.50% of the phenotypic variance, and LOD values ranged from 2.19 to 4.70 (Table 2). According to the gene annotation information of the reference genome of Brassica napus, 881, 57, 2679, 102, 526 and 72 candidate genes (QTGs) were screened from QTLs of the corresponding characters, respectively (Table S3).
Transcriptome differences between R and S lines under aluminum stress
After filtering out low-quality reads and adapter sequences, a total of 30.24 Gb of clean data was obtained. The Q30 base percentage was at least 92.69% and 80.71–86.53% of reads could be mapped to the reference genome of Brassica napus, and 77.70–83.38% were unique (Table S4). As shown in Fig. 3A and B, 8945 DEGs were obtained by comparing RCK with SCK. The number of up-regulated DEGs (4023, 45.0%) was less than that of down-regulated DEGs (4922, 55.0%). There were 5114 DEGs detected in RT vs. RCK and 4652 DEGs in ST vs. SCK. Of the 5114 DEGs in the R line, 2634 (51.5%) were up-regulated and 2480 (48.5%) were down-regulated. The 4652 DEGs of the S line included 2719 (58.4%) up-regulated genes and 1933 (41.6%) down-regulated genes. In the case of RT vs. ST, 3935 DEGs were identified including 2564 (65.2%) up-regulated genes and 1371 (34.8%) down-regulated genes. In addition, there were 1748, 1269, 849 and 3450 unique DEGs in RT vs. RCK, ST vs. SCK, RT vs. ST, RCK vs. SCK, respectively, and 78 DEGs were shared between R and S lines.
Metabolomic analysis of R and S lines under aluminum stress
Three repetitions were conducted for each treatment in the metabolic analysis to ensure the reliability of the experiment. According to PCA, the first PC (PC1) and the second PC (PC2) explained 23.4% and 21.7% of the variation, respectively, and there was an obvious separation between the control and treated samples of the R and S lines (Fig. 4A). A total of 3127 differentially accumulated metabolites (DAMs) were detected, including lipids, amino acids and their derivatives, alkaloids, carbohydrates, organic acids, terpenes, coenzymes, nucleotides and derivatives, flavonoids, alcohols, lignin and coumarins, aldehydes, phenolics and ethers (Table S5). In RT vs. RCK, there were 292 DAMs (147 up-regulated and 145 down-regulated), 353 in ST vs. SCK (276 up-regulated and 77 down-regulated), 204 in RT vs. ST (136 up-regulated and 68 down-regulated) and 117 in RCK vs. SCK, (43 up-regulated and 74 down-regulated) (Fig. 4B). After removing the DAMs with similar change trends of RT vs. RCK and ST vs. SCK, the remaining 457 metabolites were considered to be related to aluminum stress (Table S6).
Hub genes identified by integrating QTGs, DEGs and DAMs
A total of 3186 QTGs were integrated with DEGs screened by transcriptome sequencing to eliminate genes with non-significant differential expression. Based on the significant difference in QTGs expression between RT vs ST and RCK vs. SCK, or RT vs. RCK and ST vs. SCK, 372 candidate genes related to aluminum stress were detected (Table S7). Overall, the comparison of QTGs and DEGs further validated the reliability of the candidate genes and helped us mine key genes. Next, we conducted a joint screening of 372 DEGs and 457 DAMs to understand the mechanism underlying the response of rapeseed root to aluminum stress and establish the related network (|R|≥ 0.95, p ≤ 0.01). Subsequently, the correlation analysis showed that 138 hub genes exhibited strong positive or negative correlations with 30 important metabolites (14 in the positive mode, 16 in the negative mode) (Fig. 5).
As shown in Fig. 6, 138 hub genes were identified within the confidence interval of 17 QTLs, and the number of genes in each QTL interval ranged from 2 to 38. Among the 138 genes, there were 109 key genes related to RRD, followed by 48 related to RRL, 26 to RRT, and 3 key genes to RRV. There were two key genes related to RSA and RDW, and some genes were associated with two or more traits.
Functional analysis of hub genes and metabolites
The 138 hub genes were aligned by BLASTP search for sequences homologous to Arabidopsis thaliana and divided into twelve categories according to their functions (Fig. 7, Table S8). Specifically, there were seven genes related to cell wall modification, 16 to growth and development, 11 to hormone signaling pathways, 9 to protein hydrolysis, 5 to redox reactions, 7 to signal transduction, 14 to stress reactions, 9 to synthesis of secondary metabolites, 11 to transcriptional regulation, 12 to transport proteins and 20 genes to carbohydrate and lipid metabolism. In addition, there were 17 genes encoding unknown functional proteins.
The thirty candidate metabolites could be divided into ten categories: carbohydrates, lipids, amino acids and their derivatives, nucleotides and derivatives, alcohols, phenols, lignin and coumarins, aldehydes, alkaloids and coenzymes. Among them, there were six metabolites related to the synthesis of secondary metabolites, two for the metabolism of alkaloids and one associated with coenzyme synthesis. There were also 21 metabolites associated with carbohydrate, lipid, amino acid and nucleotide metabolism (Fig. 8).
Metabolic pathways associated with hub genes and metabolites
KEGG metabolic pathway analysis showed that hub genes and metabolites related to aluminum stress in rapeseed roots were mainly enriched in the metabolism of lipids, carbohydrates and secondary metabolites (Fig. 9).
Lipid metabolism
Lipids are the main components that comprise the membrane and cytoplasm of organisms and can be synthesized, stored and utilized by organisms as nutrients. Five hub genes and five differential metabolites were identified as related to lipid metabolism (Fig. 9A): BnaA05g06830D (aldehyde decarbonylase, CER1), BnaA03g24880D (phospholipase, PLD), BnaA05g25220D (GDSL-motif lipase 4, DGT), BnaC02g29610D (hydroperoxide dehydratase, AOS) and BnaA05g09340D (sterol 22-desaturase, CYP710A). In the S line, CER1 was upregulated 1.12-fold and PLD was downregulated 1.95-fold, while in the R line, CERI was upregulated 4.16-fold and PLD was downregulated 2.90-fold under aluminum toxicity. CER1 was associated with cutin, suberine and wax biosynthesis (ko00073), and the production of stem epicuticular wax, while PLD was involved in glycerophospholipid metabolism (ko00564) and played a role in membrane lipid modulation. Both AOS and CYP710A are members of the cytochrome P450 family, the former functioning as an allelic oxide synthase, while the latter plays an important role in growth and development, stress responses and pathogenesis. In addition, neg_6427 (PGD2) and neg_14145 (15-deoxy-Δ12,14-PGJ2) were involved in arachidonic acid metabolism(ko00590), and pos_9557 (13(S)-HOTrE) was involved in α-linolenic acid metabolism (ko00592). The main unsaturated fatty acids are α-linolenic acid and arachidonic acid. In the S line, pos_9557 was significantly increased 1.38-fold and neg_14145 was increased 1.33-fold; neg_6427 decreased by about 15 times in the S line exposed to Al3+ but increased by nine times in the R line.
Carbohydrate metabolism
Carbohydrates are the main energy supplying substances in living cells and have important functions in regulating cell activity. Twelve hub genes and one candidate metabolite were identified in the carbohydrate metabolism pathway (Fig. 9B). Three of these genes, chitinase (CHI), 1,4-β-D-xylan synthase (DXS) and UDP-arabinose 4-epimerase (UXE), participate in amino sugar and nucleotide sugar metabolism (ko00520). Other genes such as citrate synthase (CS), isocitrate lyase (aceA) and (S)-2-hydroxy-acid oxidase (HAO) are involved in glyoxylate and dicarboxylate metabolism (ko00630), which could facilitate the accumulation of osmotic substances to initiate osmoregulation and ROS clearance. Pectinesterase (BnaA05g04610D) and D-xylonolactone (neg_4897) were found, and they are in the pentose and glucuronate interconversions pathway (ko00040). The remaining genes, INPP5B (inositol polyphosphate 5-phosphatase), BLGX (β-glucosidase) and PPC (phosphoenolpyruvate carboxylase) are associated with phosphatidylinositol signaling (ko04070), starch and sucrose metabolism (ko00500) and pyruvate metabolism (ko00620), respectively. Among the DEGs related to carbohydrate metabolism, most of the genes in the S line exhibited greater changes than those in the R line, indicating that the S line was more sensitive to aluminum toxicity than the R line.
Metabolism of secondary metabolites
Plant secondary metabolites are a class of products synthesized by plants during growth and development, and containing substances responsible for cell growth, reproduction and aging. Eight hub genes and six metabolites were identified in the metabolism of secondary metabolites (Fig. 9C). Both BnaC01g04250D and BnaC02g35080D encoded scopoletin glucosyltransferase (TOGT1), and BnaA05g19630D encoded sinapate 1-glucosyltransferase (BRT1). These three genes and one secondary metabolite, neg_5425 (5-hydroxyferulate), were involved in phenylpropanoid biosynthesis (ko00940). BnaA05g18910D encoded geranylgeranyl diphosphate synthase (FDPS) and BnaA05g10890D encoded ent-kaurenoic acid monooxygenase (KAO); these two genes participated in terpenoid backbone biosynthesis (ko00900) and diterpenoid biosynthesis (ko00904), respectively. BnaA05g18910D regulated the accumulation of O-β-D-xylosylzeatin (neg_9752) related to zeatin biosynthesis. BnaC02g25030D encoded tropinone reductase I (TR1), BnaA03g24900D encoded magnesium dechelatase (SGR), and BnaC02g27320D encoded bifunctional dethiobiotin synthetase (BIO), which were associated with tropane, piperidine and pyridine alkaloid biosynthesis (ko00960), porphyrin and chlorophyll metabolism (ko00860), and biotin metabolism (ko00780). The secondary metabolites, neg_6371 (dihydrophaseic acid), neg_8205 (psilocybin), pos_9549 (perillyl aldehyde) and pos_1100 (NAD+) were found to be involved in carotenoid biosynthesis (ko00906), indole alkaloid biosynthesis (ko00901), limonene and pinene degradation (ko00903) and oxidative phosphorylation (ko00190), respectively.
Confirmation of candidate gene expression using qRT- PCR
To verify the accuracy and reproducibility of the transcriptome analysis, six candidate DEGs were selected for qRT-PCR analysis (Fig. 10A). The expression of these six genes was consistent with the RNA-Seq results in the R and S lines, and the Spearman correlation coefficient was 0.916 (R2 = 0.948) (Fig. 10B). These results demonstrated the reliability of the RNA-sequencing results.
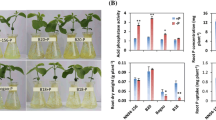
Results of qRT-PCR of candidate genes. A qRT-PCR of candidate genes. The error line represents the standard error of three technical repeats. Statistical significance was determined by LSD test. ns means not significantly different (p > 0.05); * means significantly different (p < 0.05). B Linear regression curve between qRT-PCR data and RNA-seq data. ** represents highly significant difference (p < 0.01)
Physiological indices of R and S lines under aluminum exposure
As shown in Table 3, the aluminum ion content in the roots of S line was higher than in R line. The level of POD, CAT, SOD and SP in roots of both S and R lines was significantly increased under aluminum toxicity stress (p < 0.01), but the increase in S line was less than that of R line. Consequently, the content of MDA in S line was about three times that in R line exposed to aluminum stress. This indicated that aluminum stress was more damaging to the cell membranes of S line roots.
Discussion
A combination of QTL localization and transcriptome analysis had been used previously to identify potential candidate genes for abiotic stress tolerance in crops (Marino et al. 2009; Wang et al. 2017). There were a large number of candidate genes in the QTL localization interval. Similarly, transcriptome analysis could identify thousands of DEGs associated with the target phenotypic traits, but these were still difficult to screen for key candidate genes related to target traits. Metabolomics could be used for qualitative and quantitative analysis of metabolites in specific tissues or cells of plants in a certain time and space. In this study, 138 key candidate genes related to aluminum stress in the roots of Brassica napus were identified by combining DEGs, QTGs and DAMs. Among these candidates, some are major genes that respond to aluminum stress, while the others might be chain reaction genes (minor genes). Significantly, the number of genes in each QTL interval ranged from 2 to 38 (Fig. 6). Some QTLs controlling RRL, RRD and RRT were simultaneously distributed on chromosome A05, and it was speculated that there might be major QTLs related to aluminum toxicity tolerance on that chromosome. QTL fine mapping is often considered to be one of the most important means to narrow the range of candidate genes; however, fine positioning usually requires a large crop population with heavy workload and high cost. Therefore, it is more cost-effective to combine DEGs, QTGs and DAMs to describe candidate genes associated with complex traits. We also found that some genes could affect two or more traits simultaneously (Fig. 9), which was in agreement with reports that co-mapping of QTLs for correlated traits might result from either tight linkage of multiple genes (Sandhu et al. 2001) or single gene pleiotropy (Xiao et al. 1996). The co-mapping of QTLs can help breeders identifying favorable alleles for multiple traits simultaneously in marker-assisted breeding (Liu et al. 2016).
There are two main mechanisms involved in response to aluminum stress, the aluminum exclusion mechanism and the aluminum tolerance mechanism (Singh et al. 2017), and transporters are involved in both mechanisms (Chen et al. 2022). Plants secrete organic acid anions such as malic acid through their roots, which react with Al3+ to form non-toxic complexes to resist aluminum stress (Yang et al. 2013). Tonoplast dicarboxylate transporter (TDT) is an important malic acid transporter that plays an important role in plant resistance to aluminum stress (Liu et al. 2017a, 2017b; Medeiros et al. 2017). In this study, expression of the gene BnaC02g33350D encoding TDT protein was found to be up-regulated in both the R and S lines (Table S8). Members of the ATP binding box (ABC) protein family responsible for the transfer of a variety of molecules across the plasma membrane by hydrolysis of ATP was also shown to be involved in the mechanism of aluminum resistance in plants (Ezaki et al. 2015). The ABC transporter genes, BnaA05g07200D and BnaA05g23360D, were also identified in this study. The ABC transporter OsALS1 in rice worked synergistically with the plasma membrane Al-uptake transporter, OsNrat1, to transfer aluminum ions from root cell walls into the cytoplasm where they were sequestered in vacuoles to protect plants from damage (Huang et al. 2012). Aluminum exposure upregulated lipid transporters, BnaA03g25130D, BnaA03g215150D, copper transporters, BnaA03g25640D, transmembrane proteins, BnaA05g11040D, and ammonium transporters, BnaA09g00320D. These genes could regulate and maintain cell homeostasis by influencing the transport of lipids, metal ions and penetrants.
Aluminum toxicity caused by acid soil induced reactive oxygen species (ROS), which in turn increased lipid peroxidation and protein oxidation in plants, restricted ion transport and altered membrane fluidity (Awasthi et al. 2019); however, Al also activated the oxidative stress genes, peroxidase, catalase, glutathione-S-transferase in plants which eliminated ROS (Chauhan et al. 2021). It was observed that SOD, POD and CAT activities were higher in the roots of R and S lines under Al stress (Table 3), but the increase in antioxidant enzyme activity in R line was greater than that in S line. MDA is an accepted indicator of membrane lipid peroxidation (Shahnaz et al. 2011) and the MDA content in R line was much lower than in S line exposed to Al stress, suggesting that increased antioxidant enzyme activity in the R line led to the decrease in lipid peroxidation. We also determined the expression of the redox reaction genes encoding glutathione-S-transferase (BnaA05g12560D and BnaA03g26130D) and two genes encoding cytochrome P450 (BnaA05g04960D and BnaA05g09340D), and all were upregulated in R and S lines in response to Al; but, the R line had higher expression levels than the S line (Table S8). RRTF1 (redox responsive transcription factor 1) retarded programmed cell death induced by salt stress through ROS inhibition (Soliman and Meyer 2019), and we found similar affects in R line exposed to aluminum ions. Therefore, the genes involved in redox reactions might also be important in alleviating Al3+ toxicity in plants.
Co-expression analysis of DEGs and DAMs is an effective way to identify the main metabolic pathways and key regulatory factors, influencing plant response mechanisms to stress. Plant hormones play an important role in stress alleviation (Liu et al. 2022), and the three metabolic pathways involved in the Al3+stress response in this study were mostly located upstream of genes for plant hormone synthesis (Fig. 9).The expression of the MtN21 transporter family protein, BnaC02g24380D, was upregulated in S line, but was not significantly different in R line. MtN21 is related to auxin transport, suggesting that Al3+ could interfere with polar auxin transport by up-regulating auxin transporter expression, leading to auxin accumulation in roots that inhibits root growth. During the biosynthesis of brassinosteroids (BRs), the extracellular domain of BRI1 binds BRs resulting in phosphorylation and activation of the kinase domain, which induces BR responses (Wei and Li 2016). Overproduction of BRs can inhibit root growth by altering the normal cell cycle processes in root meristem, retarding plant growth and development (González-García et al. 2011). BKI1 is a BRI1 kinase inhibitor. In this study, BnaC02g28720D, which encodes BKI1, was upregulated in the R line, while BRI1 encoded by BnaC02g34410D was downregulated; there was no significant difference in the S line. These results indicated that R line plants could prevent BR inhibition of root growth by inhibiting the BR signaling pathway. This might have been reflected in the differences in root length of R and S lines under Al3+ stress (Table 1). Application of exogenous salicylic acid increased soybean resistance to aluminum (Liu et al. 2017a, 2017b). In this study, salicylate glucose transferase 1, which was encoded by BnaA05g03580D, was significantly upregulated in both R and S lines. Thus, the complex network of Al3+-induced plant hormone changes supports the hypothesis that plant hormones are involved in the resistance to Al3+-induced stress damage in rapeseed plants.
Some transcription factors are involved in adaptive stress responses, activating or inhibiting the expression of stress-related genes to alter a plant’s ability to adapt to stressful environments (Zhong et al. 2012). In this study, the transcription factors NAC, MYB, ERF and DREB played important roles in rapeseed response to Al3+stress (Table S8). Both NAC035 (BnaC02g34540D) and MYB67 (BnaA05g26450D) were significantly upregulated in R line exposed to Al3+. Previous research suggested that the transcription factors NAC and MYB were associated with the regulation of lignification and secondary wall biosynthesis (Zhang et al. 2018; Zhao et al. 2020). Similarly, the gene BnaA05g04610D encoding pectinesterase was upregulated in both R and S lines under Al3+ stress conditions. Pectinesterase is important for the formation of the pectin cytoskeleton in plant cell walls (Jolie et al. 2010). This suggests that these genes might help to enhance aluminum resistance by promoting cell wall biosynthesis. In addition, DREB2B (BnaA05g27930D) was downregulated in R line but upregulated in S line in the presence of Al3+. AtDREB2A and AtDREB2B were strongly induced in roots by high-salt stress and in stems and roots by dehydration stress (Nakashima et al. 2000). However, how these transcription factors might influence aluminum stress responses needs further research.
In summary, the integration of QTL mapping, transcriptomics, and metabolomics can help in identifying the most important QTLs, allowing for rapid detection of the key genes and prime candidate metabolites. Using this method, a total of 138 hub genes and 30 candidate metabolites were screened, and three pathways of rapeseed root response to aluminum stress were predicted, including lipid metabolism, carbohydrate metabolism and secondary metabolite metabolism. Overall, at the gene and metabolite levels, the response of the S line to Al3+ stress was stronger than that of the R line. Our results provide much-needed practical guidance in understanding the molecular mechanisms of root responses to aluminum stress, and the collection of R and S lines affords valuable germplasm resources and genotypes for further improving aluminum tolerance in rapeseed.
Data availability
The additional files are available in the Figshare repository, https://doi.org/10.6084/m9.figshare.22756904. The raw sequence data are available in the NCBI Sequence Read Archive (SRA) repository (https://dataview.ncbi.nlm.nih.gov/object/PRJNA967236), and the accession number is PRJNA967236. Data will be made publicly accessible after publication of the manuscript.
Abbreviations
- QTL:
-
Quantitative trait locus
- RNA-seq:
-
RNA-sequence
- Al:
-
Aluminum
- RIL:
-
Recombinant inbred lines
- QTGs:
-
Quantitative trait genes
- DEGs:
-
Differentially expressed genes
- DAMs:
-
Differentially accumulated metabolites
- RRL:
-
Relative root length
- RRD:
-
Relative root average diameter
- RSA:
-
Relative root surface area
- RRV:
-
Relative root volume
- RRT:
-
Relative root tips
- RDW:
-
Relative dry weight
- POD:
-
Peroxidase
- CAT:
-
Catalase
- SOD:
-
Superoxide dismutase
- MDA:
-
Malondialdehyde
- SP:
-
Soluble proteins
- TBA:
-
Thiobarbituric acid
- UPLC-Q-TOF/MS:
-
Ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry
References
Awasthi JP, Saha B, Panigrahi J, Yanase E, Koyama H, Panda SK (2019) Redox balance, metabolic fingerprint and physiological characterization in contrasting North East Indian rice for aluminum stress tolerance. Sci Rep 9(1):8681. https://doi.org/10.1038/s41598-019-45158-3
Awasthi JP, Kusunoki K, Saha B, Kobayashi Y, Koyama H, Panda SK (2021) Comparative RNA-Seq analysis of the root revealed transcriptional regulation system for aluminum tolerance in contrasting indica rice of North East India. Protoplasma 258(3):517–528. https://doi.org/10.1007/s00709-020-01581-2
Bian M, Zhou M, Sun D, Li C (2013) Molecular approaches unravel the mechanism of acid soil tolerance in plants. Crop J 1(2):91–104. https://doi.org/10.1016/j.cj.2013.08.002
Chauhan DK, Yadav V, Vaculík M, Gassmann W, Pike S, Arif N, Singh VP, Deshmukh R, Sahi S, Tripathi DK (2021) Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit Rev Biotechnol 41(5):715–730. https://doi.org/10.1080/07388551.2021.1874282
Chen L, Wu Q, He W, He T, Wu Q, Miao Y (2019) Combined de novo transcriptome and metabolome analysis of common bean response to Fusarium oxysporum f. sp. phaseoli infection. Int J Mol Sci 20(24):6278. https://doi.org/10.3390/ijms20246278
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools–an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Chen S, Qi X, Feng J, Chen H, Qin Z, Wang H, Deng Y (2022) Biochemistry and transcriptome analyses reveal key genes and pathways involved in high-aluminum stress response and tolerance in hydrangea sepals. Plant Physiol Biochem 185:268–278. https://doi.org/10.1016/j.plaphy.2022.06.008
Doncheva S, Amenós M, Poschenrieder C, Barceló J (2005) Root cell patterning: a primary target for aluminium toxicity in maize. J Exp Bot 56(414):1213–1220. https://doi.org/10.1093/jxb/eri115
Ezaki B, Takahashi K, Utsumi K, Higashi A (2015) A half-type AvABCG1 transporter derived from Andropogon virginicus L. confers aluminum tolerance. Environ Exp Bot 118:21–31. https://doi.org/10.1016/j.envexpbot.2015.05.005
Farokhzadeh S, Fakheri BA, Mahdi Nezhad N, Tahmasebi S, Mirsoleimani A (2020) Genetic dissection of spike-related traits in wheat (Triticum aestivum L.) under aluminum stress. Genet Resour Crop Evol 67(5):1221–1243. https://doi.org/10.1007/s10722-020-00907-6
Gao H, Ye S, Wu J, Wang L, Wang R, Lei W, Meng L, Yuan F, Zhou Q, Cui C (2021) Genome-wide association analysis of aluminum tolerance related traits in rapeseed (Brassica napus L.) during germination. Genet Resour Crop Evol 68(1):335–357. https://doi.org/10.1007/s10722-020-00989-2
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327(5967):812–818. https://doi.org/10.1126/science.1185383
González-García M, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138(5):849–859. https://doi.org/10.1242/dev.057331
Han Q, Zhu Q, Shen Y, Lee M, Lübberstedt T, Zhao G (2022) QTL mapping low-temperature germination ability in the maize IBM Syn10 DH population. Plants (Basel) 11(2):214. https://doi.org/10.3390/plants11020214
Harrell F Jr (2022) Hmisc: Harrell miscellaneous. R package version., 4.7–2. https://CRAN.Rproject.org/package=Hmisc
Huang CF, Yamaji N, Chen Z, Ma JF (2012) A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J 69(5):857–867. https://doi.org/10.1111/j.1365-313X.2011.04837.x
Huang D, Gong Z, Chen X, Wang H, Tan R, Mao Y (2021) Transcriptomic responses to aluminum stress in tea plant leaves. Sci Rep 11(1):5800. https://doi.org/10.1038/s41598-021-85393-1
Jahan N, Javed MA, Khan A, Manan FA, Tabassum B (2021) Genetic architecture of Al3+ toxicity tolerance in rice F2:3 populations determined through QTL mapping. Ecotoxicology 30(5):794–805. https://doi.org/10.1007/s10646-021-02413-6
Jiang X, Yang X, Zhang F, Yang T, Yang C, He F, Gao T, Wang C, Yang Q, Wang Z, Kang J (2022) Combining QTL mapping and RNA-Seq unravels candidate genes for Alfalfa (Medicago sativa L.) leaf development. BMC Plant Biol 22(1):485. https://doi.org/10.1186/s12870-022-03864-7
Jolie RP, Duvetter T, Van Loey AM, Hendrickx ME (2010) Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohyd Res 345(18):2583–2595. https://doi.org/10.1016/j.carres.2010.10.002
Kochian LV, Piñeros MA, Hoekenga OA (2005) The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274(1):175–195. https://doi.org/10.1007/s11104-004-1158-7
Kochian LV, Piñeros MA, Liu J, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 66:571–598. https://doi.org/10.1146/annurev-arplant-043014-114822
Kopec PM, Mikolajczyk K, Jajor E, Perek A, Nowakowska J, Obermeier C, Chawla HS, Korbas M, Bartkowiak-Broda I, Karlowski WM (2021) Local duplication of TIR-NBS-LRR gene marks clubroot resistance in Brassica napus cv. Tosca. Front Plant Sci 12:639631. https://doi.org/10.3389/fpls.2021.639631
Lei L, Zheng H, Bi Y, Yang L, Liu H, Wang J, Sun J, Zhao H, Li X, Li J, Lai Y, Zou D (2020) Identification of a major QTL and candidate gene analysis of salt tolerance at the bud burst stage in rice (Oryza sativa L.) using QTL-Seq and RNA-Seq. Rice 13(1):55. https://doi.org/10.1186/s12284-020-00416-1
Li Y, Ye H, Song L, Vuong TD, Song Q, Zhao L, Shannon JG, Li Y, Nguyen HT (2021) Identification and characterization of novel QTL conferring internal detoxification of aluminium in soybean. J Exp Bot 72(13):4993–5009. https://doi.org/10.1093/jxb/erab168
Liu Z, Li H, Fan X, Huang W, Yang J, Li C, Wen Z, Li Y, Guan R, Guo Y, Chang R, Wang D, Wang S, Qiu LJ (2016) Phenotypic characterization and genetic dissection of growth period traits in soybean (Glycine max) using association mapping. Plos One 11(7):e158602. https://doi.org/10.1371/journal.pone.0158602
Liu N, Song F, Zhu X, You J, Yang Z, Li X (2017a) Salicylic acid alleviates aluminum toxicity in soybean roots through modulation of reactive oxygen species metabolism. Front Chem. https://doi.org/10.3389/fchem.2017.00096
Liu R, Li B, Qin G, Zhang Z, Tian S (2017b) Identification and functional characterization of a tonoplast dicarboxylate transporter in tomato (Solanum lycopersicum). Front Plant Sci 8:186. https://doi.org/10.3389/fpls.2017.00186
Liu H, Zhu R, Shu K, Lv W, Wang S, Wang C (2022) Aluminum stress signaling, response, and adaptive mechanisms in plants. Plant Signal Behav 17(1):2057060. https://doi.org/10.1080/15592324.2022.2057060
Lou HQ, Fan W, Jin JF, Xu JM, Chen WW, Yang JL, Zheng SJ (2020) A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ 43(2):463–478. https://doi.org/10.1111/pce.13676
Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6(6):273–278. https://doi.org/10.1016/S1360-1385(01)01961-6
Ma X, Zhao F, Su K, Lin H, Guo Y (2022) Discovery of cold-resistance genes in Vitis amurensis using bud-based quantitative trait locus mapping and RNA-seq. BMC Genomics 23(1):551. https://doi.org/10.1186/s12864-022-08788-y
Mackay TFC, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10(8):565–577. https://doi.org/10.1038/nrg2612
Marciano D, Toledo Ramos F, Neiva Alvim M, Ronaldo Magalhaes J, Giovanni Costa França M (2010) Nitric oxide reduces the stress effects of aluminum on the process of germination and early root growth of rice. J Plant Nutr Soil Sci 173(6):885–891. https://doi.org/10.1002/jpln.200900312
Marino R, Ponnaiah M, Krajewski P, Frova C, Gianfranceschi L, Pè ME, Sari-Gorla M (2009) Addressing drought tolerance in maize by transcriptional profiling and mapping. Mol Genet Genomics 281(2):163–179. https://doi.org/10.1007/s00438-008-0401-y
McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol 35(1):89–99. https://doi.org/10.1023/A:1005711431474
Medeiros DB, Barros KA, Barros JAS, Omena-Garcia RP, Arrivault S, Sanglard LMVP, Detmann KC, Silva WB, Daloso DM, DaMatta FM, Nunes-Nesi A, Fernie AR, Araújo WL (2017) Impaired malate and fumarate accumulation due to the mutation of the tonoplast dicarboxylate transporter has little effects on stomatal behavior. Plant Physiol 175(3):1068–1081. https://doi.org/10.1104/pp.17.00971
Meng L, Yang Y, Ma Z, Jiang J, Zhang X, Chen Z, Cui G, Yin X (2022) Integrated physiological, transcriptomic and metabolomic analysis of the response of Trifolium pratense L. to Pb toxicity. J Hazard Mater 436:129128. https://doi.org/10.1016/j.jhazmat.2022.129128
Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol 42(4):657–665. https://doi.org/10.1023/A:1006321900483
Qin M, Li H, Zhao N, Zhang Y, Zhang B, Liang F, Zuo K, Guo N, Tao S, Liu X, Huang Z, Xu A (2022) Integrated genomics, QTL mapping, and co-expression analyses identifying candidates of low-temperature tolerance in Brassica napus L. Ind Crop Prod 187:115437. https://doi.org/10.1016/j.indcrop.2022.115437
Sandhu D, Champoux JA, Bondareva SN, Gill KS (2001) Identification and physical localization of useful genes and markers to a major gene-rich region on wheat group 1S chromosomes. Genetics 157(4):1735–1747. https://doi.org/10.1093/genetics/157.4.1735
Sedmak JJ, Grossberg SE (1977) A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem 79(1):544–552. https://doi.org/10.1016/0003-2697(77)90428-6
Shahnaz G, Shekoofeh E, Kourosh D, Moohamadbagher B (2011) Interactive effects of Silicon and Aluminum on the malondialdehyde (MDA), proline, protein and phenolic compounds in Borago officinalis L. J Med Plants Res 5(24):5818–5827
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Singh S, Tripathi DK, Singh S, Sharma S, Dubey NK, Chauhan DK, Vaculík M (2017) Toxicity of aluminium on various levels of plant cells and organism: a review. Environ Exp Bot 137:177–193. https://doi.org/10.1016/j.envexpbot.2017.01.005
Slovak R, Ogura T, Satbhai SB, Ristova D, Busch W (2016) Genetic control of root growth: from genes to networks. Ann Bot Lond 117(1):9–24. https://doi.org/10.1093/aob/mcv160
Soliman ERS, Meyer P (2019) Responsiveness and adaptation to salt stress of the redox-responsive transcription factor 1 (RRTF1) gene are controlled by its promoter. Mol Biotechnol 61(4):254–260. https://doi.org/10.1007/s12033-019-00155-9
Tracy SR, Nagel KA, Postma JA, Fassbender H, Wasson A, Watt M (2020) Crop improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci 25(1):105–118. https://doi.org/10.1016/j.tplants.2019.10.015
Waidmann S, Sarkel E, Kleine-Vehn J (2020) Same, but different: growth responses of primary and lateral roots. J Exp Bot 71(8):2397–2411. https://doi.org/10.1093/jxb/eraa027
Wang S, Cao M, Ma X, Chen W, Zhao J, Sun C, Tan L, Liu F (2017) Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front Plant Sci 8:1427. https://doi.org/10.3389/fpls.2017.01427
Wang P, Dong Y, Zhu L, Hao Z, Hu L, Hu X, Wang G, Cheng T, Shi J, Chen J (2021a) The role of γ-aminobutyric acid in aluminum stress tolerance in a woody plant, liriodendron chinense × tulipifera. Hortic Res Engl 8(1):80. https://doi.org/10.1038/s41438-021-00517-y
Wang R, Wang L, Lei W, Wu J, Shi H, Li C, Tang Z, Li J, Zhou Q, Cui C (2021b) Screening candidate genes related to aluminum toxicity stress at germination stage via RNA-seq and QTL mapping in Brassica napus L. Acta Agron Sin 47(12):2407–2422. https://doi.org/10.3724/SP.J.1006.2021.04231
Wei Z, Li J (2016) Brassinosteroids regulate root growth, development, and symbiosis. Mol Plant 9(1):86–100. https://doi.org/10.1016/j.molp.2015.12.003
Wright KJ, Baligar VC, Wright SF (1987) Estimation of phytotoxic aluminum in soil solution using three spectrophotometric methods. Soil Sci 144:233–236
Wu J, Yuan F, Meng L, Li C, Shi H, Bai Y, Wu X, Li J, Zhou Q, Cui C (2022) QTL mapping and candidate genes screening of photosynthesis-related traits in Brassica napus L. during seedling stage under aluminum stress. Acta Agron Sin 48(11):2749–2764. https://doi.org/10.3724/SP.J.1006.2022.14225
Xiao J, Li J, Yuan L, Tanksley SD (1996) Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet 92(2):230–244. https://doi.org/10.1007/BF00223380
Xiao D, Li X, Zhou Y, Wei L, Keovongkod C, He H, Zhan J, Wang A, He L (2021) Transcriptome analysis reveals significant difference in gene expression and pathways between two peanut cultivars under Al stress. Gene 781:145535. https://doi.org/10.1016/j.gene.2021.145535
Yang L, Qi Y, Jiang H, Chen L, Kessler MR (2013) Roles of organic acid anion secretion in aluminium tolerance of higher plants. Biomed Res Int 2013:173682. https://doi.org/10.1155/2013/173682
Yang J, Fan W, Zheng S (2019) Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J Zhejiang Univ Sci B 20(6):513–527. https://doi.org/10.1631/jzus.B1900188
Ye S, Cui C, Gao H, Lei W, Wang L, Wang R, Chen L, Qu C, Tang Z, Li J, Zhou Q (2019) QTL identification for fatty acid content in Brassica napus using the high density SNP genetic map. Sci Agric Sin 52(21):3733–3747. https://doi.org/10.3864/j.issn.0578-1752.2019.21.002
Zhang J, Huang G, Zou D, Yan J, Li Y, Hu S, Li X (2018) The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol 217(2):625–640. https://doi.org/10.1111/nph.14864
Zhao Y, Zhou M, Xu K, Li J, Li S, Zhang S, Yang X (2019) Integrated transcriptomics and metabolomics analyses provide insights into cold stress response in wheat. Crop J 7(6):857–866. https://doi.org/10.1016/j.cj.2019.09.002
Zhao K, Cheng Z, Guo Q, Yao W, Liu H, Zhou B, Jiang T (2020) Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Front Plant Sci 11:571881. https://doi.org/10.3389/fpls.2020.571881
Zhong H, Guo Q, Chen L, Ren F, Wang Q, Zheng Y, Li X (2012) Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep 31(11):1991–2003. https://doi.org/10.1007/s00299-012-1311-3
Acknowledgements
We are grateful to all the colleagues in our laboratory, and thank Chongqing Engineering Research Center for providing the seeds of Brassica napus.
Funding
This research was supported by grants from the National Key Research and Development Plan (2018YFD0100500) and Chongqing Science and Technology Commission (cstc2019jscx-msxmX0383).
Author information
Authors and Affiliations
Contributions
CC and QYZ conceived the study. CYL, HSS, LX and MLX conducted the experiments. CYL wrote the original manuscript. XRW, MYN, QYZ and CC helped to revise the manuscript. HSS, YSB and JQG collected samples and measured physiological parameters. All authors have read and agreed to the published version of the manuscript. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Additional information
Communicated by Benjamin Stich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, C., Shi, H., Xu, L. et al. Combining transcriptomics and metabolomics to identify key response genes for aluminum toxicity in the root system of Brassica napus L. seedlings. Theor Appl Genet 136, 169 (2023). https://doi.org/10.1007/s00122-023-04412-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04412-z